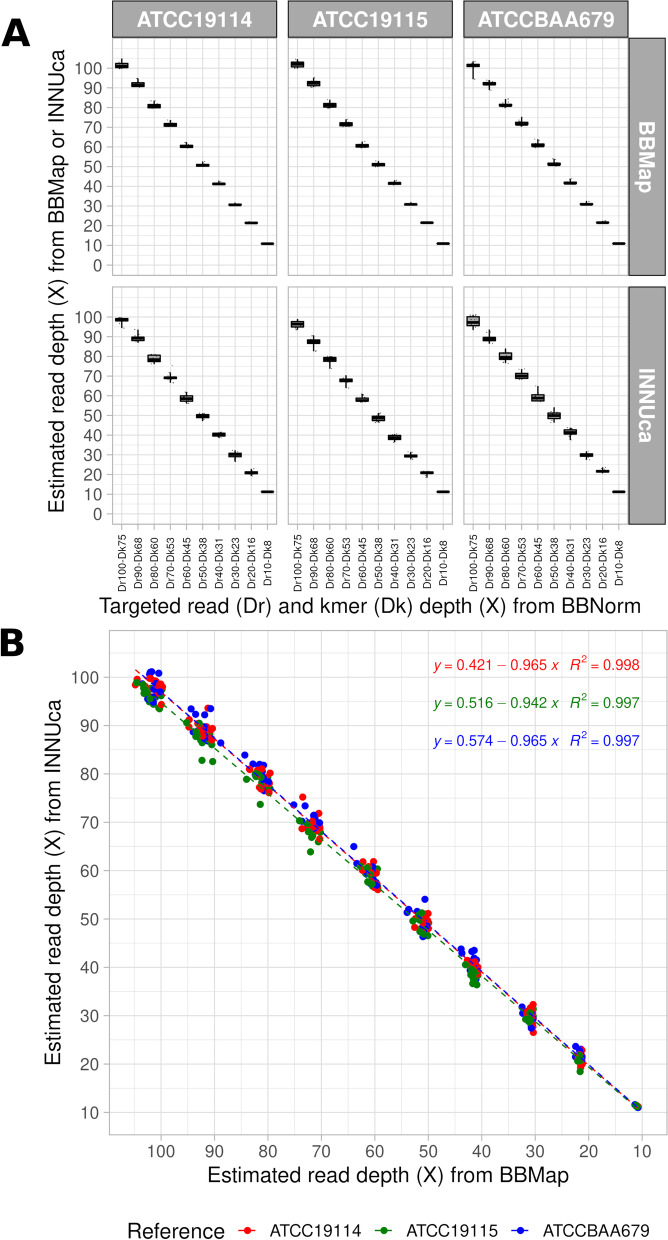
Fig. 3.
Boxplot-based distributions of targeted read (Dr) and kmer (Dk) depth (X) from BBNorm downsampling (read length R = 150 and kmer size K = 30) of Listeria monocytogenes paired-end reads from reference genomes ATCC19114, ATCC19115 and ATCCBAA679 (n = 420) according to estimated read depth (X) from BBMap (version February 13, 2020) or INNUca (version 4.2.2) with constant high read breadth of coverage (99.34 ± 0.07%) (A) and linear correlations between read depth of downsampled paired-end reads (n = 420) estimated with BBMap or INNUca for each reference genome (B)