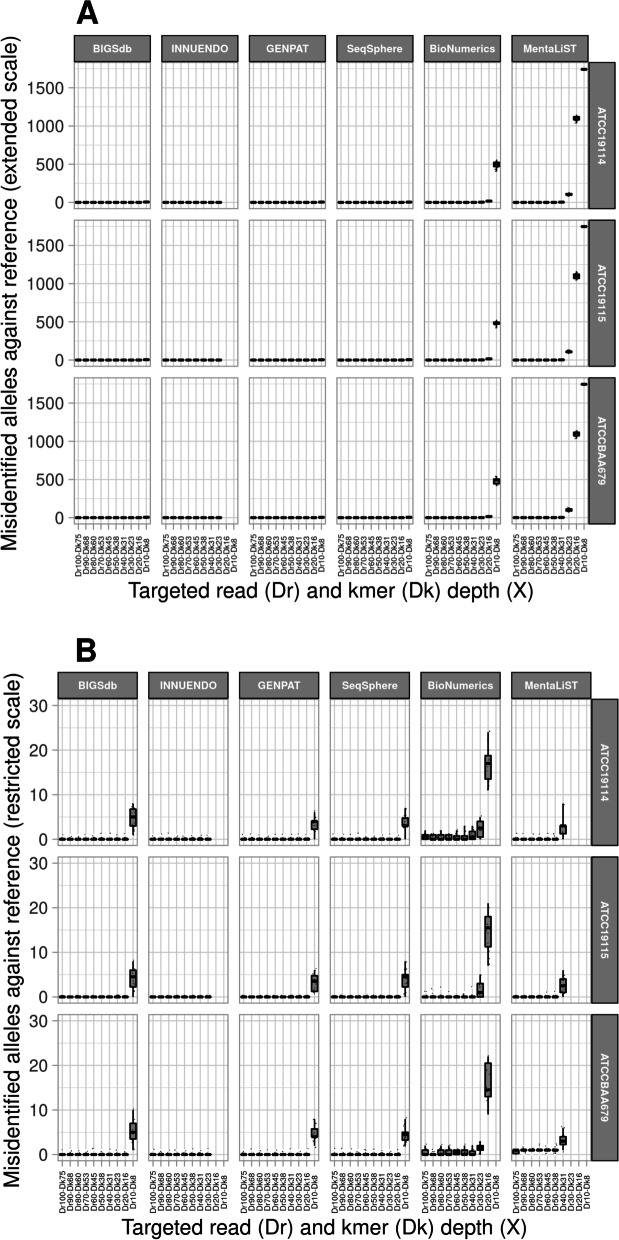
Fig. 6.
Box-plots representing the impact of downsampled paired-end reads (i.e. 2x150bp) of Listeria monocytogenes on misidentified alleles against reference at extended (A) or restricted (B) scales, according to reference genomes (i.e. ATCC19114, ATCC19115 and ATCCBAA679) and cgMLST workflows including BIGSdb (n = 420), INNUENDO (n = 336), GENPAT (n = 420), SeqSphere (n = 420), BioNumerics (n = 420) and MentaLiST (n = 420). The targeted read depth (Dr: 10X, 20X, 30X, 40X, 50X, 60X, 70X, 80X, 90X and 100X) were prepared according to kmer depth (Dk): 8X, 15X, 23X, 30X, 38X, 45X, 52X, 60X, 67X, 75X) setting of BBNorm (read length R = 150 and kmer size K = 30). Because of internal firewall, the INNUca assembler integrated into the cgMLST workflow INNUENDO cannot not perform assemblies of paired-end reads with read depth of coverage of 20X (n = 42) and 10X (n = 42)