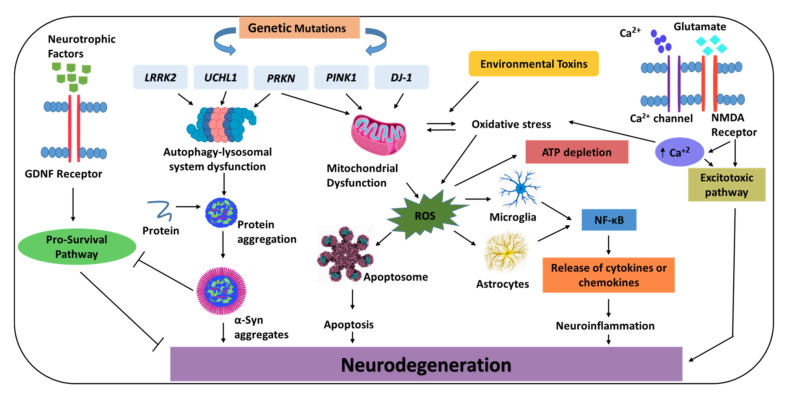
Figure 1.
Molecular pathways involved in the pathophysiology of Parkinson’s disease (PD). Common pathogenic mechanisms in PD, including genetic mutation, defective protein clearance, mitochondrial dysfunction, loss of trophic factors, alterations of intracellular Ca2+ homeostasis, and neuroinflammation, are illustrated. Enhanced signaling pathways are indicated by solid arrows, and suppressed pathways are by blocked arrows. GDNF: glial cell-derived neurotrophic factor, LRRK2: leucine-rich repeat kinase 2, UCHL1: ubiquitin C-terminal hydrolase L1, PRKN: parkin, PINK1: phosphatase and tensin homolog-induced kinase 1, DJ-1: Daisuke-Junko-1, α-Syn: alpha-synuclein, ROS: reactive oxygen species, TNF-α: tumor necrosis factor-alpha, IL-1β: interleukin-1β, IL-6: interleukin-6, NMDA: N-methyl-D-aspartate.