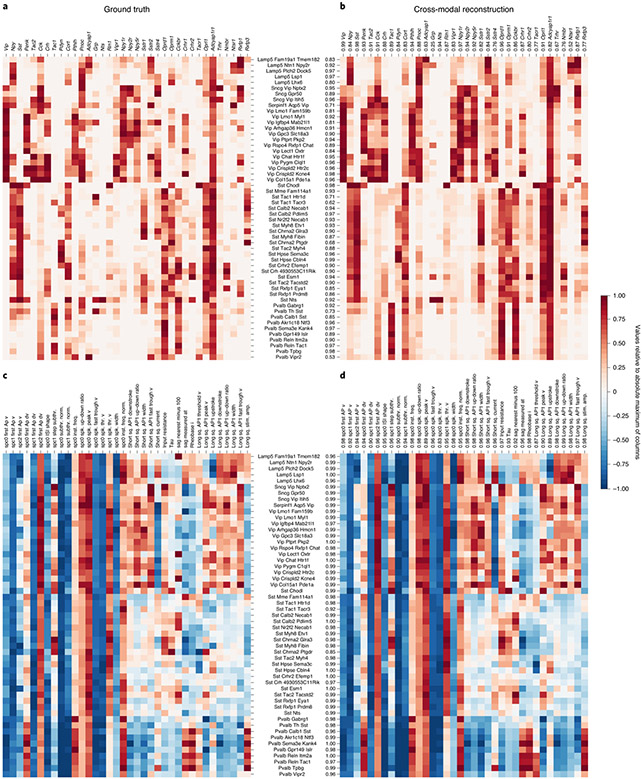
Fig. 2 ∣. Cross-modal reconstructions capture cell-type-specific gene expression patterns and electrophysiological features.
a, Gene expression levels averaged over samples reference taxonomy cell types, normalized per gene by the maximum value of each column. b, The cell-type specificity of different genes is captured well by cross-modal prediction of gene expression profiles from electrophysiological features (Pearson’s r = 0.89 ± 0.10, mean ± s.d. over cell types). c, A subset of electrophysiological features pooled by cell types shows analogous cell-type specificity. d, Cross-modal reconstructions of the electrophysiology features from gene expression profiles match the measured electrophysiology features (Pearson’s r = 0.98 ± 0.02, mean ± s.d. over cell types). Cell-type and feature-wise fidelity of reconstructions are quantified with Pearson’s r for each row and column in b and d as compared to ground truth in a and c.