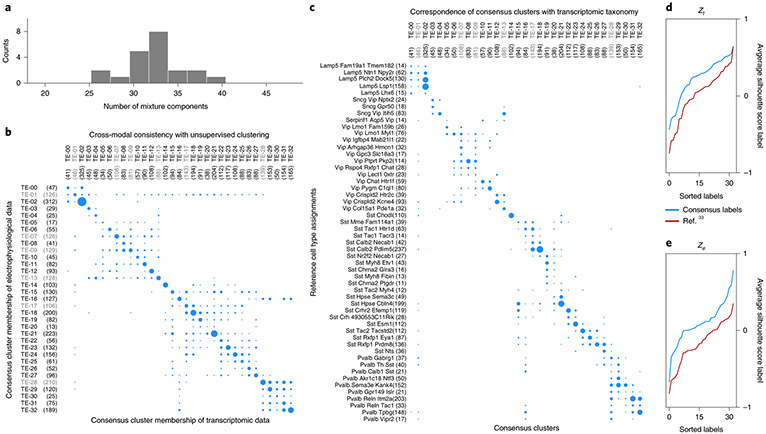
Fig. 3 ∣. Deriving a consensus cell-type clustering.
a, Unsupervised clustering using Gaussian Mixtures on the coordinated representation zt and BIC-based model selection suggests 33.0 consensus clusters (32.19 ± 3.16, mean ± s.d.). b, Contingency matrix for cluster assignments based on independent, unsupervised clustering of the transcriptomic and electrophysiology representations shows that the clusters are highly consistent. c, Contingency matrix for the leaf node cell-type labels of the reference hierarchy compared with unsupervised cluster assignments show that these unsupervised clusters have substantial overlap with known transcriptomic cell types. The number of cells for each label are indicated within parentheses next to the label, and area of the dots is proportional to number cells in the scatter plots of b and c. d, Consensus clusters are compared with an equal number of cell classes obtained by merging the reference hierarchical taxonomy, using silhouette analysis based on coupled representations. Average per-cluster silhouette values for test cells are larger for consensus cluster labels. Clusters for which the silhouette score is less than zero in d and e are grayed out in panels b and c.