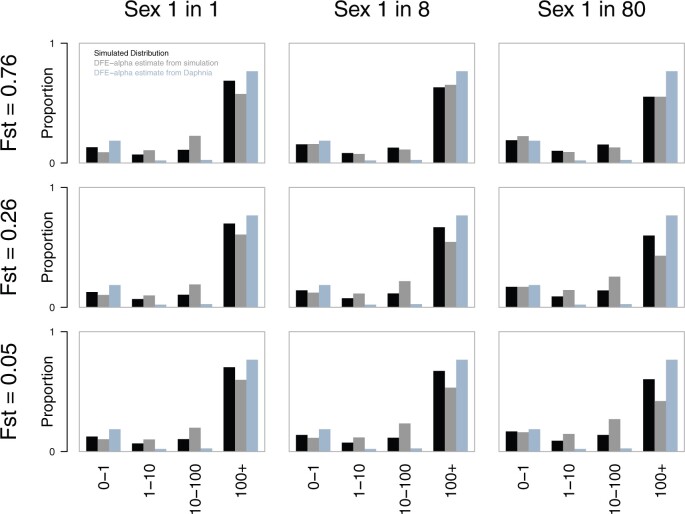
Fig. 7.
Simulated (black) and estimated (gray, as shown for the controls in fig. 6) DFE proportions for three levels of frequency of sexual reproduction and population subdivision (FST), respectively. Using SLiM 2.0, we simulated coding DNA sequence-like data for one diploid individual sampled from each deme of a 36-deme structured population and inferred the DFE and . Mutation and recombination rates were selected to match those of D. magna (after scaling, see Materials and Methods), and selection coefficients were chosen to reflect those estimated for D. melanogaster, which generally shows a high proportion of adaptive substitutions and a small proportion of weakly deleterious mutations. Combinations of population subdivision (and thus migration) and sexual reproduction rates were explored, bracketing credible values for D. magna. Consistent with theoretical expectations, strong population subdivision (low migration) reduced the efficacy of natural selection, and rare sex reduced the efficacy of positive selection, at least in part through the constraint which arises from link sites. The estimated proportions of DFE were broadly consistent with estimates of observed in the simulation (table 1; see supplementary table 3, Supplementary Material online for observed values). These results suggest the effects of population structuring and asexuality (which are unaccounted for by DFE-alpha or asymptotic MK) are in the general direction of but still insufficient to completely obscure Drosophila-like levels of adaptive protein substation.