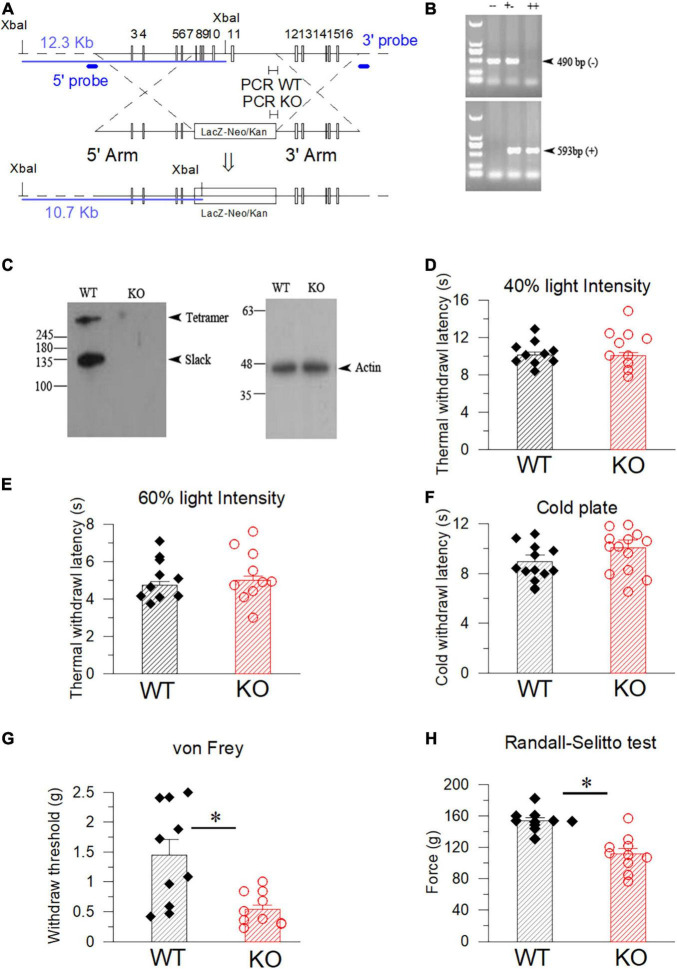
FIGURE 1.
The strategy of constructing and validating the Slack–/– mice and examination of its pain behavior. (A) The KCNT1 targeting construct in which the DNA sequence of genome locus containing 7–11 exon of the Slack channel gene was replaced by IRES-Lacz-Neo/Kan Cassette. The nuclease cleavage site, the Southern Blot probe, and expected bands of the WT and Slack–/– mice were shown. The locus of the PCR fragment for identification of the WT mice and Slack KO mice at the indicated position. (B) PCR verification of the mice genotypes using the mice tail tissue. The PCR product shows a band of 593bp in electrophoresis for WT mice, while the amplicon for the Slack–/– mice is 490 bp. PCR products of the heterozygote mice contain both fragments. (C) Western Blot verification of missing the Slack channel protein using the protein samples extracted from the spinal cord tissues of the WT and Slack–/– mice. (D,E) WT mice and Slack–/– mice exhibit no difference in paw withdrawal time under light-induced hot planar test with 40% light intensity (WT: 10.2 ± 0.3 s, n = 10; Slack–/–: 10.1 ± 0.3 s, n = 10. Student T-test: P = 0.49) and 60% light intensity (WT: 4.8 ± 0.2 s, n = 10; Slack–/–: 5.0 ± 0.2 s, n = 10; P = 0.74). (F) WT mice and Slack–/– mice exhibit no difference in paw withdrawal time under dry ice induced cold planar test (WT: 9 ± 0.5 s, n = 13, Slack–/–: 10 ± 0.6 s, n = 13; P = 0.67). (G) Mechanical thresholds of the WT mice (1.4 ± 0.3 g, n = 10) and Slack–/– mice (0.5 ± 0.1 g, n = 10) show a statistically significant difference (von Frey test, student T-test: P = 0.035, *indicates P < 0.05). (H) Enhanced pain threshold in Slack–/– mice with Randall-Selitto test (WT: 154 ± 4.2 g, n = 10; Slack–/–: 112 ± 6.9 g, n = 10; student T-test: P = 0.0004, *indicates P < 0.05).