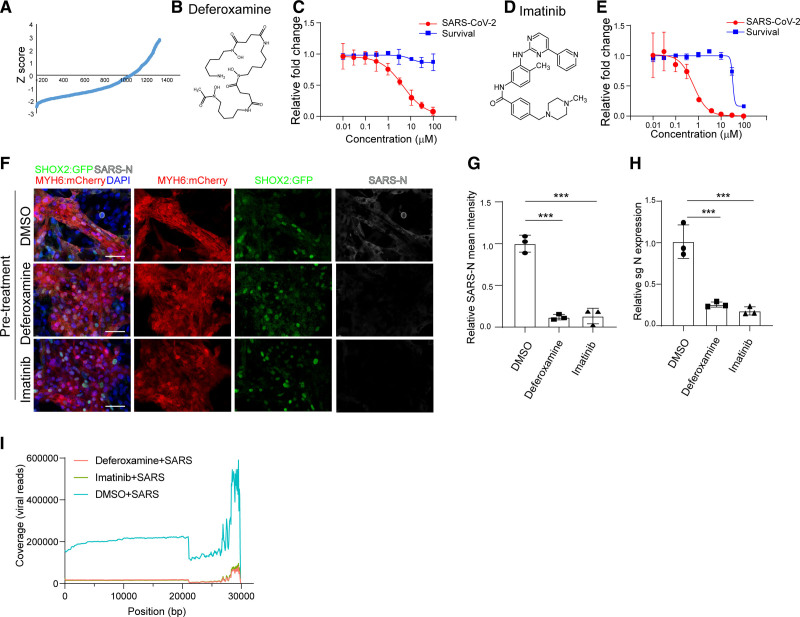
Figure 6.
A high content chemical screen to identify anti–severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) drugs using human embryonic stem cell (hESC)–sinoatrial node (SAN)–like pacemaker cells. A, The primary screening result. x axis represents the compound number. y axis represents the Z score. B, Chemical structure of deferoxamine. C, Efficacy and cytotoxicity curves for deferoxamine. D, Chemical structure of imatinib. E, Efficacy and cytotoxicity curves for imatinib. F and G, Immunostaining (F) and the quantification (G) of dimethyl sulfoxide (DMSO), 50 μM deferoxamine-treated, or 10 μM imatinib-treated hESC-SAN–like pacemaker cells at 24 hours postinfection (hpi) with SARS-CoV-2 (multiplicity of infection [MOI]=0.1). Scale bar=50 μm. H, Quantitative real-time polymerase chain reaction analysis of total RNA extracted from DMSO, 50 μM deferoxamine-treated, or 10 μM imatinib-treated hESC-SAN–like pacemaker cells at 24 hpi with SARS-CoV-2 (MOI=0.1) for viral N subgenomic RNA (sgRNA). The graph represents the mean sgRNA level normalized to ACTB. I, Read coverage of the viral genome from DMSO, 50 μM deferoxamine-treated, or 10 μM imatinib-treated hESC-SAN–like pacemaker cells at 24 hpi with SARS-CoV-2 (MOI=0.1). N=3 independent experiments. Data were presented as mean±SD. DAPI, indicates 4′,6-diamidino-2-phenylindole; MYH6: mCherry, myosin heavy chain 6: mcherry; SARS-N, SARS-CoV-2 nucleocapsid; sgN, sub genomic SARS-CoV-2 nucleocapsid RNA; and SHOX2:GFP, short stature homeobox 2: green fluorescent protein. P values were calculated by 1-way ANOVA with an indicated control. ***P<0.001.