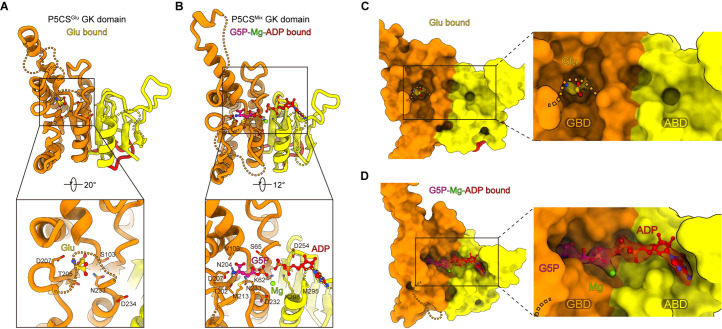
Figure 2. Conformational changes in the glutamate kinase (GK) domain-binding pocket.
(A) GK domain of the P5CSGlu filament, with glutamate shown as sticks with yellow carbons. The dashed lines represent disordered segments (residues 124–142, 211–232, and 275–297) in this model. (B) GK domain of the P5CSMix filament, with G5P, Mg+, and ADP shown as sticks with pink, green, and red carbons, respectively. The dashed lines represent disordered segments (residues 128–140, 214–228, and 282–295) in this model. (C, D) GK domain model surface representation showing the conformation of the binding pocket in the P5CSGlu filament or P5CSMix filament. The cryo-electron microscopy (cryo-EM) density of binding glutamate molecule in (C), and the binding complex of G5P, Mg+, and ADP in (D). The dashed lines represent ‘open loop’ and ‘closed loop’