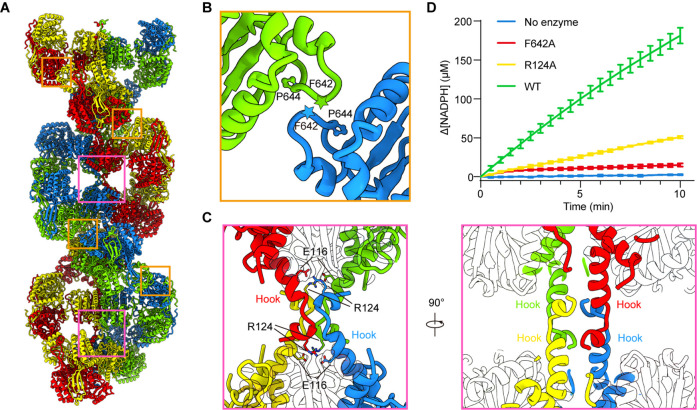
Figure 5. Assembly and interaction surfaces of the P5CS filament.
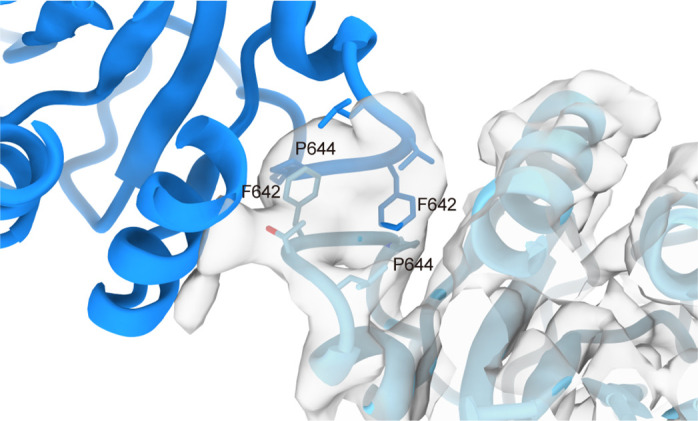
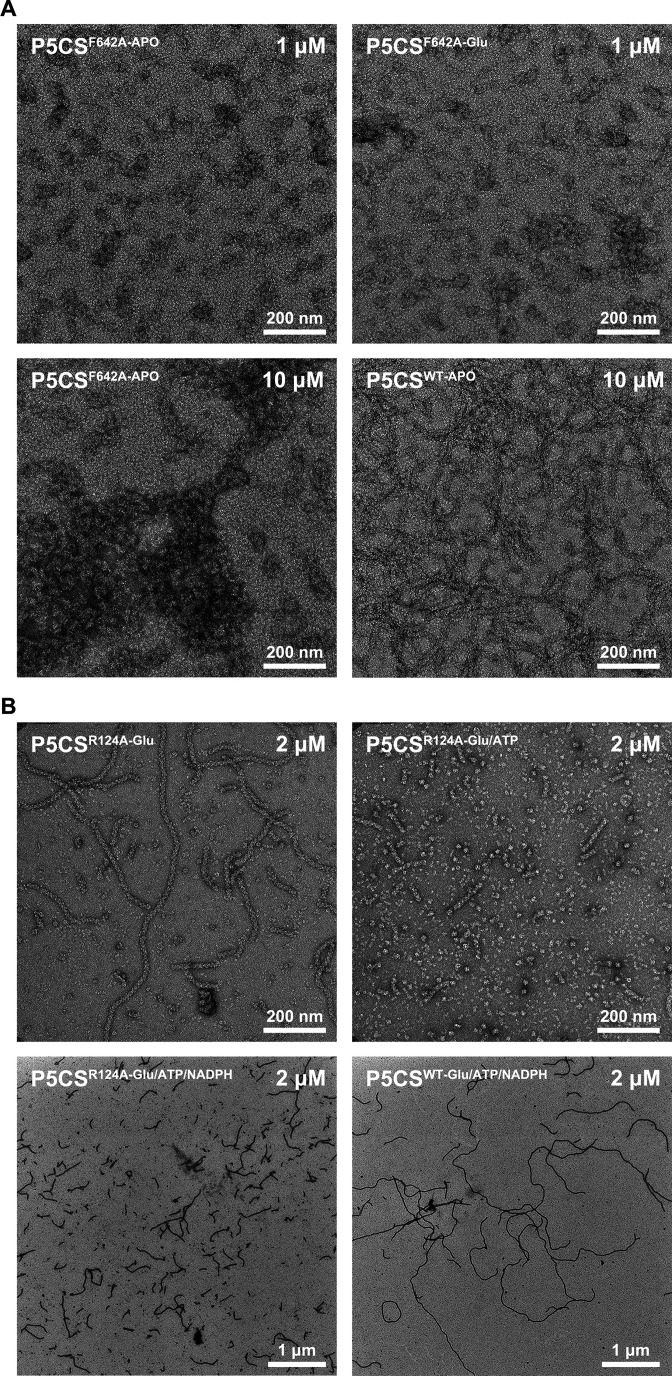
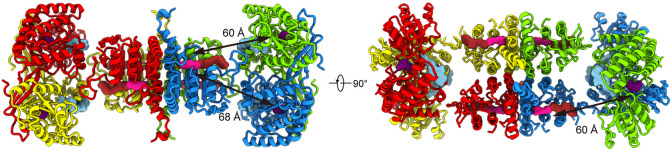
(A) P5CS filament assembly interface, the four P5CS protomers in one layer are colored in red, yellow, blue, and green. (B) Interaction between two adjacent γ-glutamyl phosphate reductase (GPR) domain dimers, residues F642 located at loop that interacts with P644 from another neighboring GPR domain dimer. (C) Model for hook structure interaction. (D) Enzyme activity analysis to examine P5CS wild-type or mutant proteins. All of the experiments were replicated three times (n = 3, mean ± SD).
Figure 5—figure supplement 1. The interface of adjacent γ-glutamyl phosphate reductase (GPR) domain dimers.