FIGURE 4.
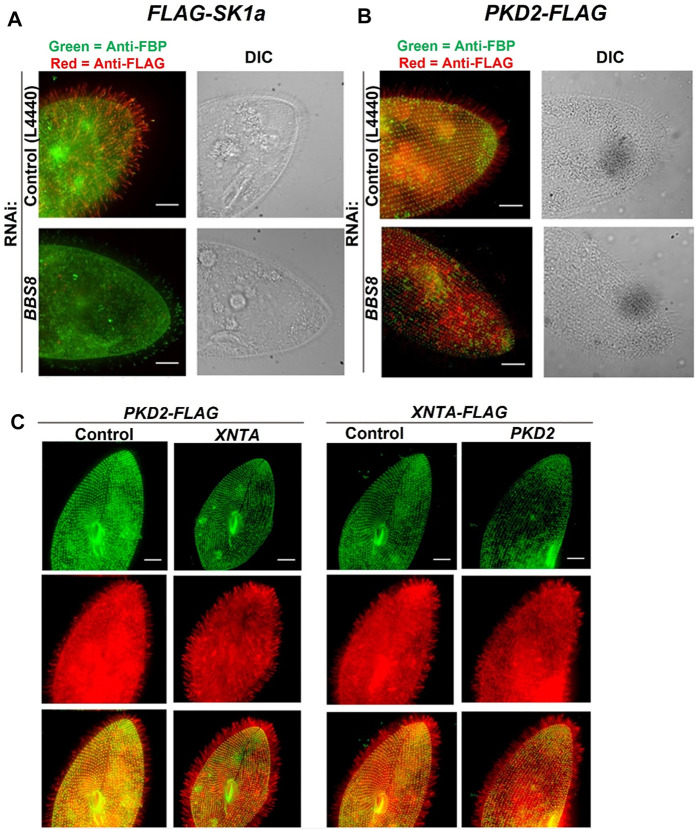
Immunofluorescence and RNAi allow us to follow the location of channels and other proteins in the cell. (A) immunofluorescence allowed us to localize the Sk1a channel (FLAG-Sk1a, red) to mainly the cilia of Paramecium (green staining shows the GPI-anchored folate binding protein, FBP). Upon feeding BBS8 RNAi, the FLAG-SK1a disappears from the cilia, showing the dependence of this channel on the BBSome for trafficking. The GPI-anchored FBP remains undisturbed. Similarly (B) cells expressing PKD2-FLAG show the Pkd2 channel at the cell surface and in the cilia (red; basal bodies are green, stained with anti-Tetrahymena centrin) and after feeding the cells RNAi bacteria to deplete BBS8, the PKD2-FLAG channel is absent from the cilia. We have also used expressing cells and RNAi to examine the dependence of interacting proteins on one another for trafficking (C) Cells expressing PKD2-FLAG depleted in XNTA show no change in the localization of the Pkd2 channel and visa versa; cells expressing XNTA-FLAG depleted in PKD2 show no change in the localization of the XntA protein. Scale bars represent 15 µm (A) and (B) from Valentine et al., 2012 reproduced with changes with permission using Creative Commons CC BY license (C) reproduced with permissions from Valentine et al., 2019 with permission using Creative Commons CC BY license.