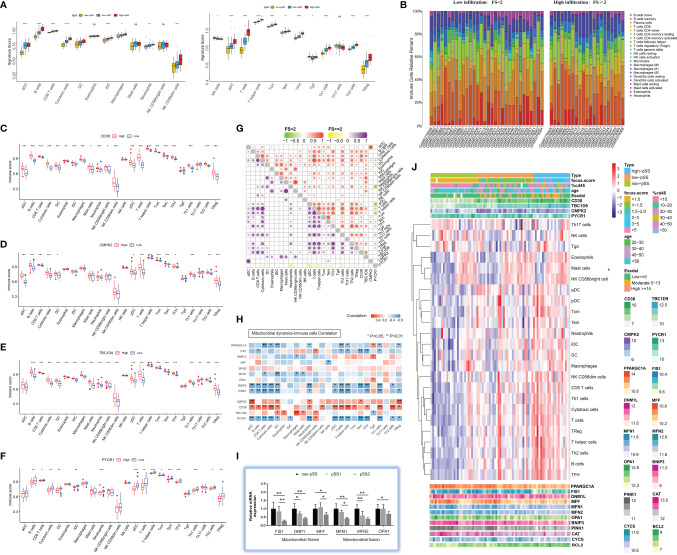
Figure 4.
Characterization of immune infiltration in salivary glands in pSS. (A) Boxplot of the normalized signature score based on single-sample gene set enrichment analysis (ssGSEA) in the validation cohort. The yellow boxplot represents non-pSS patients, the blue boxplot represents low-infiltration pSS patients, and the red boxplot represents high-infiltration pSS patients. ***p < 0.001, **p < 0.01, *p < 0.05, ns = 1 (independent sample Kruskal–Wallis test). (B) Comparisons of immune-cell proportion between pSS-low-infiltration and pSS-high-infiltration groups in the validation cohort. (C–F) Comparison of 24 immune cell infiltration for CD38 (C), CMPK2 (D), TBC1D9 (E), and PYCR1 (F) high-expression groups versus low-expression group. (G) Spearman correlation between 4 hub genes (CD38, CMPK2, TBC1D9, and PYCR1) and 24 immune cells in pSS-low-infiltration and pSS-high-infiltration groups. (H) Correlation between interested genes (4 hub genes and mitochondrial dynamic-related genes) and 24 immune cells in pSS patients. (I) The expression profile of genes regulating mitochondrial dynamics was evaluated by quantitative real-time PCR from our own cohort. Data shown are normalized to Actin expression and are relative to expression in the non-pSS (n = 3, error bars represent mean ± SD). *p < 0.05, and **p < 0.01 by Student’s t test. (J) Heatmap shows the enrichment scores of interested genes (4 hub genes and mitochondrial dynamic-related genes) and immune cells in pSS samples. Each column represents an individual patient sample, and each row represents a gene or an immune cell, ordered by unsupervised hierarchical clustering.