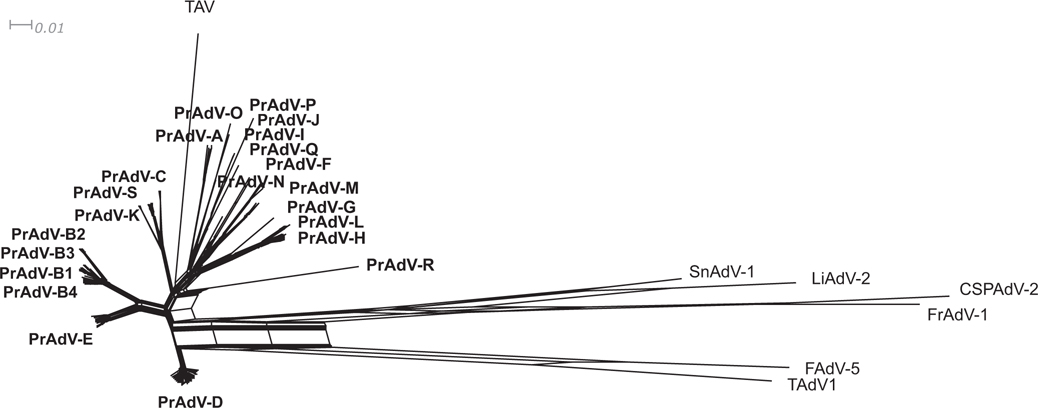
Fig. 2.
Phylogenetic network analysis of Mastadenovirus genomes. Whole genome sequences (174 total) that have been archived in GenBank were collected and analyzed. A phylogenetic network was constructed using SplitsTree software with default parameters applied to character blocks using the GTR + I + G algorithm model (http://www.splitstree.org/). These genomes represented all human and simian adenoviruses contained in the genus Mastadenovirus, along with six non-Mastadenovirus outgroup viruses and one closely related nonprimate adenovirus (treeshrew, TAV). As shown in this network, the proposed primate adenovirus species (PrAdV) segregate into 19 species or clades, with one comprising subclades (PRAdV-B1 to B4). The ingroup for the adenovirus clades is in bold for each cluster. Provided is a scale bar that represents the number of weighted splits (“bipartisan”).