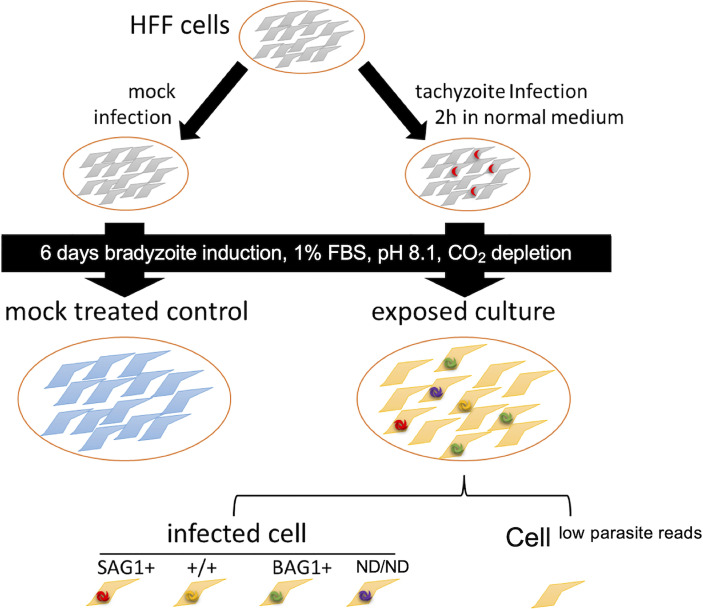
Figure 1.
Schematic diagram of the experimental design. For transcriptome analysis of parasite-infected cells, we prepared bradyzoite-infected cells (exposed culture) by inoculating HFF cells with tachyzoites, followed by the induction of bradyzoite differentiation for 6 days in bradyzoite induction medium without CO2 addition. As a control, HFFs without parasite inoculation were incubated under the same conditions for 6 days (mock-infected treated control). In the exposed culture, the cells were assigned to each category based on the mRNA molecules detected. Parasite-mapped mRNA molecules (parasite load) were used to infer the infection status, and the cells were divided into infected cells and others (cellslow parasite reads). Infected cells were further assigned to different subsets based on the presence of the canonical tachyzoite marker SAG1 and bradyzoite marker BAG1 (SAG1+: cells with T. gondii SAG1+/BAG1−, +/+: cells with both BAG1 and SAG1 markers were detected, BAG1+: cells with T. gondii SAG1−/BAG1+, ND/ND: cells with T. gondii, but both SAG1 and BAG1 markers were below the detection limit).