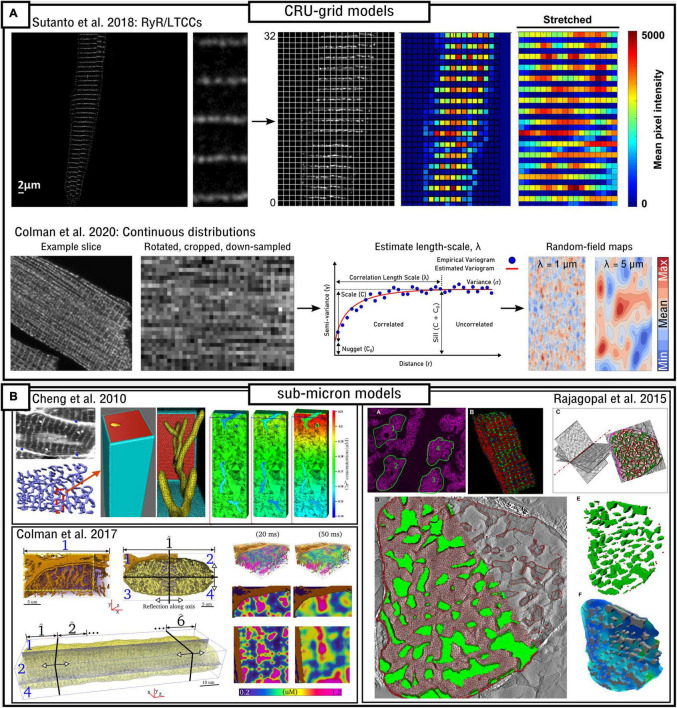
FIGURE 7.
Image-based modeling approaches. (A) Approaches for image-based modeling in idealized, CRU-grid cell models. The top panel illustrates the processing of LTCC/RyR image data so that it can be aligned and registered on an idealized grid computational model, as presented in Sutanto et al. (2018). The lower panel illustrates the method of calculating the correlation length-scale (λ) in order to generate Gaussian-random field maps which match these parameters in order to describe sub-cellular heterogeneity, as presented in Colman et al. (2020). (B) Approaches for directly incorporating imaging data into realistic, sub-micron cell models. The top-left panel demonstrates processing of ultra-structural data describing T-system structure into a mesh for simulations, and illustrates the large Ca2+ gradients that can occur under different flux-distributions, as presented in Cheng et al. (2010). Lower panel illustrates processing of data describing the T-system and SR in a portion of a cell, approaches to tesselate the cell portion into a whole-cell, and the substantial Ca2+-gradients observed in this model also, as presented in Colman et al. (2017b). Right panel illustrates the semi-automated pipeline for segmenting multiple structures, including the myofibrils and mitochondria, into a 3D structurally-detailed representation of a sarcomere, as presented in Rajagopal et al. (2015).