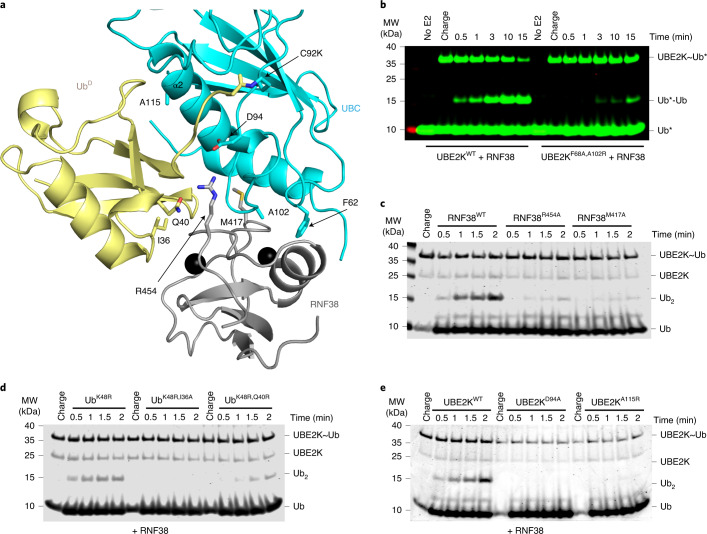
Fig. 2. Validation of the UBE2K/UbD/RING network.
a, Structure of UbD (yellow) with the C terminus covalently attached to the UBE2K (cyan) active site K92 and primed for catalysis by RNF38 RING (gray). The isopeptide bond between UBE2K K92 and UbD G76 along with key residues in the binding interface are shown as sticks. b, Nonreduced SDS–PAGE showing di-Ub formation with the RING-binding-deficient UBE2K mutant. Asterisks (*) indicate fluorescent-labeled Ub. c, Nonreduced SDS–PAGE showing di-Ub formation in the presence of RNF38 E2/UbD-binding-deficient mutants. d, Nonreduced SDS–PAGE showing the effect of UbD mutations on di-Ub formation with RNF38. e, Nonreduced SDS–PAGE of RNF38 with UBE2KD94A and UBE2KA115R in di-Ub formation. Coomassie staining was used to visualize c, d and e.