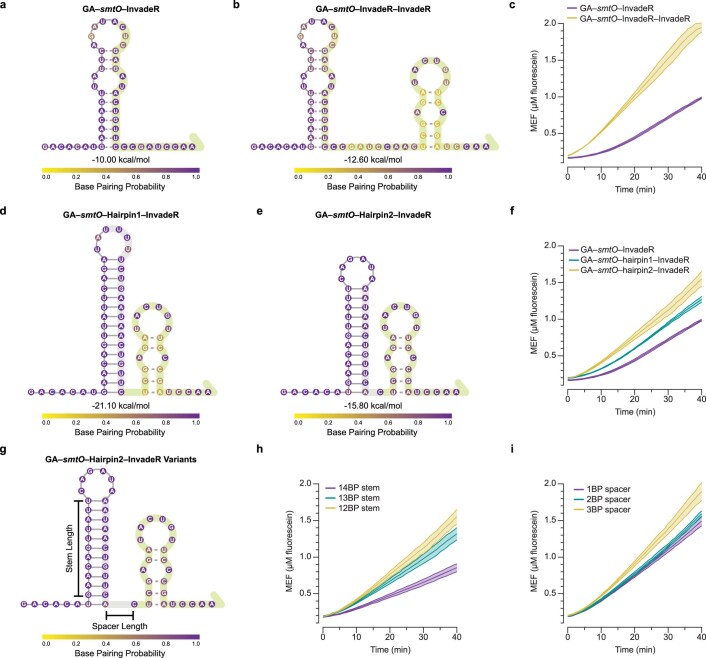
Extended Data Fig. 4. Extra nucleotides can be added between the operator and InvadeR sequence to tune the kinetics of the zinc sensor.
a, Secondary structure, minimum free energy and base pairing probabilities of GA-smtO–InvadeR predicted by NUPACK at 37 °C30. A part of the wild type smtO sequence forms a strong predicted stem-loop with InvadeR. The nucleotides highlighted in green correspond to the InvadeR sequence that strand-displaces the DNA signal gate. b, Secondary structure, minimum free energy and base pairing probabilities of GA-smtO–InvadeR–InvadeR predicted by NUPACK at 37 °C30. c, Comparison of the kinetics of the unregulated GA-smtO–InvadeR reaction and the unregulated GA-smtO–InvadeR–InvadeR reaction. d, e, NUPACK-predicted secondary structures, minimum free energies and base pairing probabilities of the two GA-smtO–Hairpin–InvadeR variants designed to sequester the smtO sequence and prevent it from binding to InvadeR. The nucleotides highlighted in grey indicate the added sequestering sequence. f, Comparison of the kinetics of the unregulated reactions of the variants shown in d and e. g, The GA-smtO–Hairpin2–InvadeR variants were built by lengthening either the stem length or the spacer between the hairpin and the InvadeR sequence. The stem length variants are built with the 0-nt spacer, and the spacer variants are built with the 12-bp stem length. h, i, Comparison of the kinetics of the unregulated reactions of the GA-smtO–Hairpin2–InvadeR variants. All data shown are n = 3 independent biological replicates each plotted as a line with raw fluorescence values standardized to MEF (μM fluorescein). Shadings indicate the average of the replicates ± standard deviation.