Fig. 3. Transcription of InvadeR can be regulated by aTFs.
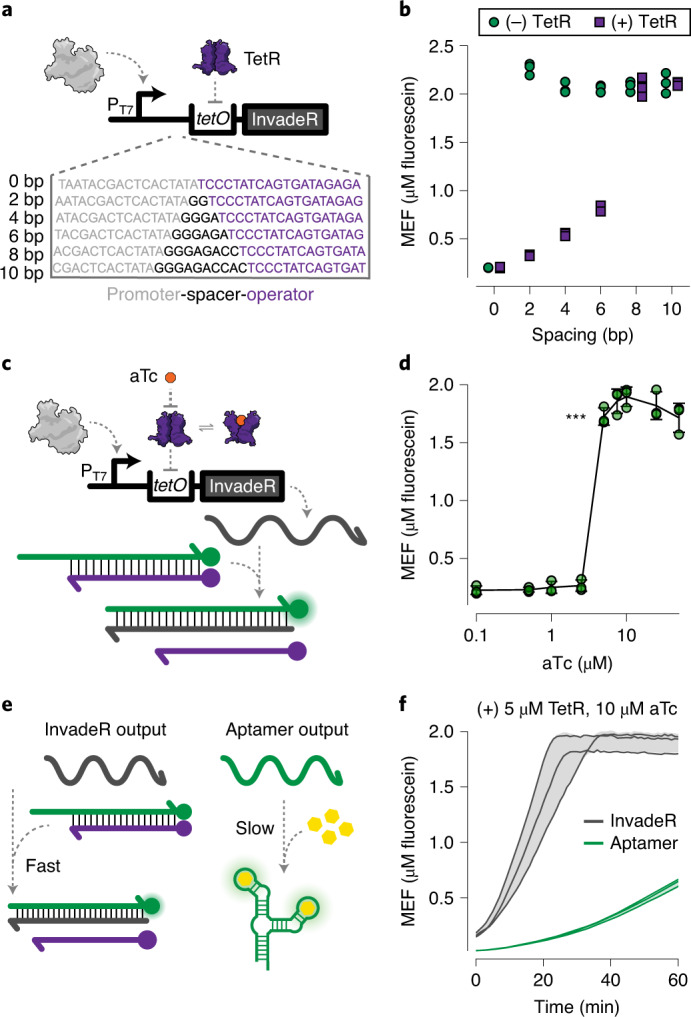
a, IVTs can be allosterically regulated with a template configured to bind a purified aTF (TetR) via an operator sequence (tetO) placed downstream of the T7 promoter. A series of spacers at 2-bp intervals was constructed to evaluate the impact of spacer length on the ability of TetR to regulate the transcription of InvadeR. b, Endpoint data (at 1 h) shown for promoter–operator spacer variants regulated (with 5 μM TetR dimer, 50 nM DNA template) and unregulated (without TetR). c, Induction of a TetR-regulated IVT reaction occurs in the presence of the cognate ligand, aTc, which binds to TetR and prevents its binding to tetO. This allows transcription to proceed, leading to fluorescence activation via TMSD. d, Dose response with aTc, measured at 1 h with 50 nM DNA template and 5 μM TetR dimer. The lowest ligand concentration at which the signal is distinguishable from the background was determined using a two-sided, heteroscedastic Student’s t-test against the no-ligand condition, and the P value range is indicated by asterisks (***P < 0.001, **P = 0.001–0.01, *P = 0.01–0.05, the exact P value for 5 µM aTc = 5.064 × 10−5). Exact P values along with degrees of freedom for all ligand concentrations tested can be found in the source data. Data for the no-ligand condition were excluded because the x axis is on the log scale and are presented in the source data. e, The speed of the TMSD output is faster than that of the RNA aptamer output. f, Comparison of fluorescence kinetics between the TetR-regulated InvadeR and aptamer outputs when induced with 10 μM aTc. All data shown are n = 3 independent biological replicates each plotted as a point (b,d) or a line (f) with raw fluorescence values standardized to MEF (μM fluorescein). Error bars (d) and shading (f) indicate the average of the replicates ± s.d.
