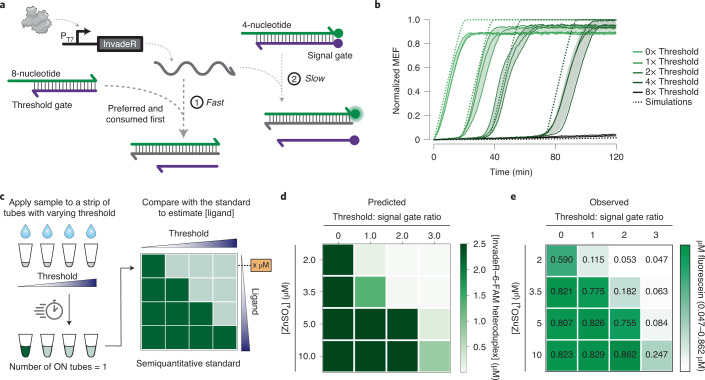
Fig. 6. Quantifying ligand concentration with a genetic ADC circuit.
a, Increasing the length of the DNA gate toehold region can be used to speed the strand invasion process. An unlabeled DNA gate with a longer toehold (eight nucleotides) can then preferentially react with InvadeR, acting as a programmable threshold. InvadeR can only strand-displace the signal gate (four-nucleotide toehold) after the threshold gate is exhausted. b, Titrating the eight-nucleotide toehold threshold gate in different ratios above a fixed signal gate concentration (0–8×) results in a time delay in fluorescence activation that can be quantitatively modeled with ODE simulations (dotted lines). All data shown for n = 3 independent biological replicates each plotted as a line. Raw fluorescence values were first standardized to MEF (μM fluorescein) and normalized to the maximum MEF among all conditions to accommodate their comparison with the simulations (See Methods for the normalization method used). Shading indicates the average of the replicates ± s.d. c, A molecular ADC circuit is made by constructing a strip of tests of the same sensor, with each test containing a different concentration of the DNA threshold gate. A higher threshold gate concentration requires a higher ligand concentration to activate fluorescence. When the same sample is applied to each tube, a user can obtain semiquantitative information about the concentration of ligand present in the sample (analog input) by identifying the series of tubes that activate (binary digital output). Here, we define a tube with its MEF value >0.5 as ‘ON’ because the visible threshold is around the indicated value. d,e, Characterization of a molecular ADC circuit for zinc using ODE simulations (d) and endpoint experimental data at 100 min (e) generated using the SmtB-regulated zinc sensor. The values on the heatmap represent the average MEF (μM fluorescein) of n = 3 independent biological replicates (see Extended Data Fig. 8 for all data).