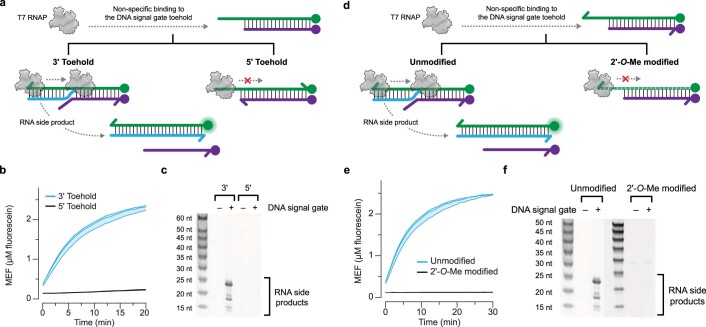
Extended Data Fig. 1. Toehold-mediated DNA strand displacement can be used to track RNA outputs with an appropriately designed DNA gate.
a, In the presence of IVT components, T7 RNAP can non-specifically bind to the toehold region of the DNA signal gate. When the toehold is on the 3’ end of the gate, this leads to transcription of unwanted RNA side products that can displace the quencher strand. This process is blocked when the toehold is on the 5’ end of the gate. b, The 3’ toehold DNA signal gate leads to fluorescence activation in the presence of T7 RNAP, while the 5’ toehold DNA signal gate does not. c, When the reaction products from b were extracted and run on an urea-polyacrylamide gel, RNA side products appear only from the 3’ toehold DNA signal gate. A negative control where no DNA signal gate is present in the reaction (–) was run alongside for both 3’ and 5’ toeholds. d, Modifying the DNA gate with 2’-O-methyl oligonucleotides prevents promoter-independent transcription by T7 RNAP. e, When the 3’ toehold DNA signal gate is modified with 2’-O-methyl oligonucleotides, no fluorescence activation is observed in the absence of a T7 RNAP-driven IVT template. f, When the reactions from e were run on an urea- polyacrylamide gel, no RNA side products were observed from the 2’-O-methyl DNA signal gate. A negative control where no DNA signal gate is present in the reaction (–) was run alongside for both unmodified and modified gates. Data shown in b and e are n = 3 independent biological replicates each plotted as a line with raw fluorescence standardized to MEF (μM fluorescein). Shading indicates the average of the replicates ± standard deviation. Data shown in c and f are a representative of n = 3 independent biological replicates. The uncropped, unprocessed gel image shown in c and f is available as Source Data.