Abstract
Background
The acute respiratory syndrome coronavirus 2 (SARS-CoV-2) disease seriously affected worldwide health. It remains an important worldwide concern as the number of patients infected with this virus and the death rate is increasing rapidly. Early diagnosis is very important to hinder the spread of the coronavirus. Therefore, this article is intended to facilitate radiologists automatically determine COVID-19 early on X-ray images. Iterative Neighborhood Component Analysis (INCA) and Iterative ReliefF (IRF) feature selection methods are applied to increase the accuracy of the performance criteria of trained deep Convolutional Neural Networks (CNN).
Materials and methods
The COVID-19 dataset consists of a total of 15153 X-ray images for 4961 patient cases. The work includes thirteen different deep CNN model architectures. Normalized data of lung X-ray image for each deep CNN mesh model are analyzed to classify disease status in the category of Normal, Viral Pneumonia and COVID-19. The performance criteria are improved by applying the INCA and IRF feature selection methods to the trained CNN in order to improve the analysis, forecasting results, make a faster and more accurate decision.
Results
Thirteen different deep CNN experiments and evaluations are successfully performed based on 80-20% of lung X-ray images for training and testing, respectively. The highest predictive values are seen in the analysis using INCA feature selection in the VGG16 network. The means of performance criteria obtained using the accuracy, sensitivity, F-score, precision, MCC, dice, Jaccard, and specificity are 99.14%, 97.98%, 99.58%, 98.80%, 97.81%, 98.83%, 97.68%, and 99.56%, respectively. This proposed study is indicated the useful application of deep CNN models to classify COVID-19 in X-ray images.
Keywords: COVID-19, Convolutional neural networks CNN, Iterative neighborhood component analysis, Iterative ReliefF, Feature selection
1. Introduction
The coronavirus is a virus that causes respiratory system infection and can be transmitted from person to person. This virus was seen in the Wuhan region of China in early December 2019 and soon spread to other countries. The name of the virus has been declared by the World Health Organization (WHO) as SARS-CoV-2. It also uses the term COVID-19 to describe the illness caused by the virus. The coronavirus, which has been mutating and spreading since August 2021, has infected approximately 203,295,170 million people worldwide, according to the official statements of the WHO, and caused approximately 4,303,515 million deaths [1]. The most common symptoms are fever (39° and above), cough, weakness, shortness of breath. Clinical features alone cannot define the diagnosis of COVID-19 in patients with early onset of symptoms. Among the nucleic acid-based tests, the Reverse Transcription Polymerase Chain Reaction (RT-PCR) test was used to confirm COVID-19 positive patients [2]. RT-PCR sometimes fails to diagnose COVID-19, and as a result, patients do not receive appropriate treatment in a timely manner. Therefore, a negative RT-PCR test does not rule out COVID-19 infection [3]. Availability of testing kits poses important problem fruitful detection of the illness. It is very important the application of deep learning and artificial intelligence based approaches for the efficient detection of disease from X-ray images. During the current COVID-19 pandemic, using such deep learning based approaches in real time, especially for rapid testing, successful implementation, and detection of disease, could potentially provide enormous benefits [[4], [5], [6], [7], [8]].
1.1. Related studies
Early detection of COVID-19 disease is very important. A certain portion of the patient population affected by this disease, the infection can cause severe organ failure and death [9]. Although X-ray imaging is widely used in hospitals in almost most countries, unfortunately there is a deficiency of specialists to analyze and interpret X-ray images in some low-resource clinics and developing countries. A number of DL methods were tried for the analysis of these images. Data mining and data analysis have identified the potential value of big data in the educational process as part of popular technologies in the information fields. Some of these studies will be discussed in this part of the study. In Ref. [10], the authors used image improving techniques such as histogram equalization. They also used gamma correction, balance contrast improvement techniques, image complement, contrast limited adaptive histogram equalization to research the effect of image improvement techniques on COVID-19 detection. In addition, the proposed U-net model was compared with the standard U-Net model for lung image. Six pre-trained CNN; ResNet101, ResNet18, InceptionV3, ResNet50, ChexNet and DenseNet201 were analyzed on flat and segmented lung CXR images. Ucar and Korkmaz [11] developed COVID-19 method named COVIDdiagnosis-Net based on Deep Bayes-SqueezeNet, deep learning network, for diagnosis of COVID-19 with the help of Bayes optimization. In Ref. [12], X-ray images of various resolutions were trained with different ConvXNets forms designed. A stacking algorithm was used to optimize the estimates. Also, they integrated a gradient-based discriminant localization to separate abnormal area of the X-ray images. In the study of Nour et al. [13], the CNN model was used as a deep feature extractor. Also, extracted deep distinguishing features were used machine learning algorithms with a Support Vector Machine (SVM), Decision Tree (DT) and k-Nearest Neighbor (k-NN). Bayesian Optimization Algorithm was used for optimize of machine learning models. Fuzzy tree transform was applied to each image, and then sample splitting was applied to these images by Tuncer et al. [14]. They used the multicore native binary pattern for feature generation and the features were selected by the iterative neighboring component feature selector method. Then, images were analyzed using several algorithms such as k-NN, SVM, DT. Ozturk et al. [15] suggested a model for early find of COVID-19 cases using X-ray images. The DarkNet model was formed as the classifier YOLO object detection system model. They worked on automated detection of COVID-19 based on combined hybrid feature selection and sample pyramid feature extraction [16]. In addition, pyramid feature generation based on samples with fused dynamic dimensions, feature selection based on ReliefF and iterative neighbor component analysis was performed. Artificial neural networks (ANN) and Deep neural networks (DNN) were used to classify the selected most informative features. Marqueset et al. [17] analyzed of the X-ray images obtained in two stages. Firstly, binary classification analysis was performed using X-ray images taken from normal and COVID-19 images. Second, multiclass results using images from normal, pneumonia and COVID-19 patients were compared. The proposed EfficientNet was used for dual and multiclass. A convolutional neural network-based Decomposition, Transmission, and Rendering (DeTraC) model were developed by Abbas et al. [18]. Class boundaries were analyzed using the class decomposition mechanism, which helps the model fit with any anomaly in the image dataset. Karaknis et al. [19] applied synthetic images to rise the restricted number of lung X-ray images. They used dual classification for normal cases, COVID-19 and multiclass classification for normal cases, pneumonia and COVID-19. Ismael et al. [20] extracted features from CXR images from a dataset with normal patient and COVID-19 images using a pre-trained ResNet50 model. In the classification models achieved the best accuracy using the linear-core SVM. Also, in Ref. [21], Zebin and Rezvy applied pre-trained EfficientNetB0, ResNet50, VGG16 networks to determine COVID-19 and concerned infection based on lung X-ray images. In addition, a productive oppositional structure was trained for the formation and growth of the COVID-19 class. Researchers have been working hard since the beginning of the pandemic to find automated COVID-19 detection systems that use DNN. A comparative of recent studies is shown in Table 1 . The data used in this study [22] were acquired from diverse sources and preprocessed. A deep CNN network was developed to enable detection of COVID-19 cases. When the dataset was small, data augmentation was used to artificially generate more samples from the same dataset instead of collecting more data. Similar to our study, a three-class classification (Normal, Viral Pneumonia and COVID-19) were used. Two datasets were used in these studies. The initial dataset include only 180 images cases of COVID-19. 25 cases of pneumocystis, SARS and Streptococcus were defined as pneumonia. The second dataset includes 8851 normal and 6012 pneumonia. As noted, 180 positive COVID-19 cases exist. In our study, more data was used than the data of the study of [22]. ResNet18 network was designed to reduce model complexity. As a result of the analysis, it was estimated with 96.73% accuracy. The results of the cascade classifiers were proposed in the study [23], which included the best detections of bacterial pneumonia images, viral (non-COVID-19) pneumonia and COVID-19 were seen for the ResNet50V2, VGG16 and DenseNet169 networks with 99.9% accuracy. However, despite the classification of four different diseases, the number of dataset is less than our study. In the study of [23], 306 X-ray images with four classes as 79 images for viral (Non-COVID-19) pneumonia, 79 normal, 69 positive COVID-19 images, and 79 bacterial pneumonia cases were used. Saha et al. [24] proposed an automated detection called EMCNet to determine COVID-19 patients. The extracted features, machine learning classifiers (SVM, Random Forest, DT, and AdaBoost) were developed for COVID-19 detection. Proposed EMCNet dataset contains 4600 images. The data set was divided into three different sets: the training set with 3220 images at the rate of 70%, the validation set with 920 images at the rate of 20%, and the test set with 60% images at the rate of 10%. Compared to other DL based systems, EMCNet outperformed with 98.91% accuracy, 98.89% F1 score and 97.82% recall. An automated method of DL-assisted using X-ray images for early detection of COVID-19 infection was proposed [25]. The 286 images in the training set contain 143 normal case and 143 COVID-19 images. In the test dataset, there are 60 normal case and 60 COVID-19 images. Classification of eight pre-trained CNN models from COVID-19 and normal cases, such as VGG16, AlexNet, GoogleNet, SqueezeNet, MobileNet-V2, ResNet34, ResNet50 and InceptionV3 networks, was studied. The best performance was achieved by ResNet34 network with 98.33% accuracy. Demir [26] used a deep LSTM architecture called Deep-Coronet to automatically determine COVID-19 cases from X-ray images. In addition, marker-controlled watershed segmentation and Sobel gradient processes were implemented to the images to increase the performance of the proposed model in the preprocessing phase. The estimation accuracy was calculated as 100% for the reason of using small dataset as indicated in the paper. In our study, 15153 data are used: 80% of this data is used for training and 20% for testing. In the study of [26], there was a total of 1061 data: 20% for testing, 80% of this data was used for training. 100% accuracy was obtained with the data set which is smaller than ours.
Table 1.
Some studies diagnosing COVID-19 Chest X-ray with CNNs.
| Model | Accuracy (%) | Classes | Type |
|---|---|---|---|
| Modified ResNet-18 [22] | 96.73 | COVID-19, Normal, Pneumonia | CXR |
| VGG16,ResNet50V2, DenseNet169 [23] | 99.9 | Bacterial pneumonia, COVID-19, Non-COVID-19 Viral | CXR |
| EMCNet [24] | 98.91 | COVID-19, Normal | CXR |
| ResNet-34 [25] | 98.33 | COVID-19, Normal | CXR |
| DeepCoroNet [26] | 1.00 | COVID-19, Normal, Pneumonia | CXR |
There are some studies in which CT images, X-ray images, or two images together are analyzed to determine the diagnosis of COVID-19 with the help of deep learning methods [6,[27], [28], [29], [30], [31], [32], [33], [34]].
1.2. Our method
In this study, a large-scale method of diagnosing COVID-19 is presented. This method is consisted of preprocessing, classification using deep CNN, classification with feature selection INCA and IRF. In the preprocessing step, the input Chest X-Ray image is converted to RGB converted Portable Network Graphics (PNG) image with a size of 299x299 and 8 bit depth and resized according to the required input size of the meshes used for classification. The features produced with the help of deep CNN are combined, and the distinctive ones are selected using INCA and IRF. One of the most preferred distance-based feature selection methods is ReliefF. It is a parametric property selector. 10-fold cross-validation is used for both feature selections. The features selected by INCA and IRF are again used as input to deep CNN classifiers.
1.3. Contributions
The main contributions of this study are given below:
-
•
Two different feature selection methods (INCA and IRF) are applied on the same dataset to increase the efficiency of classification and to reduce the validation time. It is seen that the use of these feature selection algorithms gave efficient results.
-
•
With the help of these methods, very high classification accuracies were calculated for thirteen different CNN. These results are denoted the overall success of the proposed IRF and INCA method.
-
•
In addition, when the number of features is huge, using feature selection is one of the most important steps for machine learning. To optimize this step, INCA and IRF feature selectors are used and higher performance criteria are obtained with this model then the performance criteria of unfeatured selection model.
The remainder of this study is organized as follows: In Section 2, dataset collection and preparation, feature selection and evaluation metrics are mentioned. Section 3 presents the experimental results and comparisons. Section 4 presents an extensive discussion with similar literature studies. Finally, in Section 5, the conclusions and future work are discussed.
2. Material and method
2.1. Material
RT-PCR testing for the detection of COVID-19 has a very laborious, long and complex process. Therefore, we selected an evident and publicly available dataset containing Chest X-ray images to respond the need for a fast disease diagnosis system. To obtain a private COVID-19 dataset, three different datasets, the COVID-19 Chest X-ray dataset [35], the Viral Pneumonia Chest X-ray dataset [36], the Normal Chest X-ray dataset [37] is merged. The resulting COVID-19 dataset occurs of 15153 Chest X-ray images for 4961 patient cases. The dataset includes 10192 Normal, 1345 Viral Pneumonia, and 3616 COVID-19 infection cases. In viral pneumonia samples, the diseases are caused by bacterial and non-COVID-19 viral effects. The used dataset consists of Portable Network Graphics (PNG) image files with a size of 299x299. It is transformed to RGB with 8-bit depth. The workflow of the proposed Deep CNN framework to classify the COVID-19 situation in Chest X-ray images is visually presented in Fig. 1 . First, lung X-ray images are collected and these images are labeled as three different lung X-ray images (Normal, Viral Pneumonia and COVID-19). After the dataset is labeled in three different classes, the images in the dataset are converted to RGB format. Data are divided as 80% training, 20% testing. The training dataset is analyzed separately with the help of thirteen different deep CNN. First, the classification of the extracted features as a result of the analysis is performed. Then, these features are selected and reclassified using INCA and IRF feature selection algorithms. Fig. 2 illustrates a sample of Chest X-ray images used for this study. All three datasets are updated regularly whereby hospitals around the world.
Fig. 1.
Workflow of proposed Deep CNN framework for classifying the COVID-19 status in X-Ray images.
Fig. 2.
Sample images of Normal (a), Viral Pneumonia (b), and (c) COVID-19 Chest X-rays.
The entire collections of 15153 Chest X-ray images have proper distributions to train and test the proposed model. The dataset contains three classes and a randomly selected set of images from class samples is shown in Fig. 3 . This study is carried out by the deep learning network of MATLAB R2020a with a personal computer with 16 GB RAM and 3.30 GHz processor.
Fig. 3.
Training data (a) and test data (b) used in deep learning analysis of the dataset.
2.2. Method
In this study, a three-class classification of Normal, COVID-19 and Viral Pneumonia Lung X-ray images are performed using VGG16, VGG19, Squeezenet, Shufflenet, Resnet101, Resnet50, Resnet18, GoogleNet, DarkNet53, DarkNet19, AlexNet, DenseNet201, InceptionResnet-V2 deep CNN. Feature vectors generated using thirteen pre-trained networks are shown in Table 2 below.
Table 2.
Generated feature vectors using the thirteen pretrained networks.
| Network | Layer | Length |
|---|---|---|
| VGG16 | ‘fc8′ | 1000 |
| ‘drop7′ | 4096 | |
| VGG19 | ‘fc8′ | 1000 |
| ‘drop7′ | 4096 | |
| Squeezenet | ‘conv 10′ | 1000 |
| ‘drop 9′ | 512 | |
| Shufflenet | ‘node_202′ | 1000 |
| ‘node_200′ | 544 | |
| Resnet101 | ‘fc1000′ | 1000 |
| ‘pool5′ | 2048 | |
| Resnet50 | ‘fc1000′ | 1000 |
| ‘avg_pool' | 2048 | |
| Resnet18 | ‘fc1000′ | 1000 |
| ‘pool5′ | 512 | |
| GoogleNet | ‘loss3-classifier' | 1000 |
| ‘pool5-drop_7x7_s1′ | 1024 | |
| DarkNet53 | ‘conv 53′ | 1000 |
| ‘avg1′ | 1024 | |
| DarkNet19 | ‘conv 19′ | 1000 |
| ‘avg1′ | 1024 | |
| AlexNet | ‘fc8′ | 1000 |
| ‘drop7′ | 4096 | |
| DenseNet201 | ‘fc1000′ | 1000 |
| ‘avg_pool' | 1920 | |
| InceptionResnet-V2 | ‘predictions' | 1000 |
| ‘avg_pool' | 1536 |
2.2.1. Feature selection
Feature selection is the process of evaluating the features according to the algorithm used and selecting the best k features from among the n features in the data set.
2.2.1.1. Iterative Neighborhood Component Analysis (INCA)
Neighborhood Component Analysis (NCA) is one of the weight based feature selectors used to maximize the prediction accuracy of the most preferred classification algorithms. Weights are calculated for all feature columns [38]. First, it assigns a fixed weight to all attribute columns. It usually sets the value 1. An optimizer such as ADAM, distance-based fitness function, and stochastic gradient descent (SGD) are used to update the weights. Informative and redundant weights represent larger and smaller weights, respectively. Also, NCA generates only positive weights [39]. NCA has two main problems. First, there is no classifier or an optimal feature vector for a problem. Secondly, since there is no negative weighted feature, the redundant feature is selected as elimination. The INCA feature selector is an iterative and increased model of NCA to accomplish these problems [40]. The primary purpose of the INCA feature selector is to find the optimum number of features. For this reason, an iterative error calculation process is used in INCA. A classifier is chosen as the error/loss value calculator, the feature vector with the minimum error value is chosen as the optimum feature vector. INCA selects a variant number of features for variant problems [41]. In this study, we selected the feature range from 50 to 1000. Firstly, data is normalized. We normalized 3030 sized feature vectors using min-max normalization.
| (1) |
where X is the final feature vector with a size of 3030. Therefore, the normalization process must be applied to obtain optimum results. Then, sorted indices are obtained by applying NCA to the normalized features. NCA weights are generated by using Eq. (2).
| (2) |
In order to calculate error value, k-nearest neighborhood (k-NN) is used. In this work, k-NN with Manhattan distance is used. Weights are sorted by descending with Eq. (3).
| (3) |
where endex is sorted indices with a length of 3030 and target is the actual/real outputs.
Besides, this process has high calculation complexity. To reduce this complexity, a feature range of 50 to 1000 is defined. The iterative feature selection process is defined as follows:
| (4) |
| (5) |
is selected feature vector in the iteration, defines size of the selected feature vector.
Calculated error values of :
| (6) |
10-fold cross-validation is used to calculate errors. After this calculation, the minimum error is calculated.
| (7) |
Finally, the best feature () is selected by using a calculated index value.
| (8) |
The generated is forwarded to classifiers.
2.2.2. Iterative ReliefF (IRF)
Relief is a widely used feature selection algorithm that is feature precision and finds the weights of the estimator when the output of the estimator depends on a multiclass variable. By using the relief algorithm, feature ranking is made for the sample selected from the data set, taking into account the proximity of other classes in its class and its distance from other classes. This feature ranking is run with a model with negative and positive weights, and the feature selection process is completed. The feature reduction method can be used to increase the classification ability [42].
ReliefF generates both positive and negative weights and uses the Manhattan distance to calculate the weights. ReliefF is an enhanced model of Relief. Euclidean distance is used in the relief based feature selection method. The Manhattan distance is used to construct the ReliefF weights [43].
ReliefF automatically selects the most prominent features. Therefore, the iterative ReliefF method is proposed. A loss calculator should be chosen for optimum properties. Therefore, a k-NN classification method is used as the loss value calculator. In this study, both feature selections are evaluated and an increase in performance criteria is observed as a result of the analysis.
2.3. Evaluation metrics
In order to evaluate the performance of the deep learning model, different metrics which are accuracy, specificity, sensitivity, precision, F-score, Kappa, Matthews correlation coefficient, Dice and Jaccard are applied in this study to measure the correct and/or incorrect classification of COVID-19 detected in the tested Chest X-ray images. Used evaluation metrics in this study are given in Table 3 below.
Table 3.
Evaluation metrics used in the analysis.
| (9) | |
| (10) | |
| (11) | |
| (12) | |
| (13) | |
| (14) | |
| (15) | |
| (16) | |
| (17) | |
| (18) | |
| (19) |
where, True Positive, True Negative, False Negative and False Positive are TP, TN, FN and FP, respectively. Also value of beta is 1.
3. Results
The goal of this study is to predict the COVID-19 viral infection using various deep CNN. For this purpose, three different datasets are collected. This dataset contains 3616 COVID-19, 10192 Normal and 1345 Viral Pneumonia heterogeneous Chest X-ray images. These data are analyzed in Matlab R2020a program in thirteen different deep CNN (Vgg16, Vgg19, Squeezenet, Shufflenet, Resnet101, Resnet50, Resnet18, GoogleNet, DarkNet53, DarkNet19, AlexNet, DenseNet201, and InceptionResnet-V2). Chest X-ray images are preprocessed before making predictions in deep CNN. The dataset is grayscale images. So the images are replicated three times to create an RGB image and these images are resized as needed for deep CNN. The data is divided into 80% training and 20% testing. Before analyzing the training data of deep CNN, some training parameters are adjusted. The set training options are shown in Table 4 below.
Table 4.
Parameters used in CNN training.
| Training Options | Parameter |
|---|---|
| Optimization algorithm | ‘sgdm' |
| Mini Batch Size | 64 |
| Max. Epochs | 10 |
| Initial Learn Rate | 0.0001 |
| Learn Rate Drop Factor | 0.6 |
| Learn Rate Drop Period | 20 |
After adjusting the training options, COVID-19 detection is performed on thirteen different deep CNN separately. After the training, the extracted features are analyzed separately in the INCA and IRF feature selection algorithms and the features are extracted. Again, it is given as input data to deep CNN. The workflow of the proposed deep CNN framework to classify the COVID-19 condition in Chest X-ray images is shown in Fig. 1. The performance criteria of the obtained results are shown separately in the tables. Thirteen different deep CNN classified data collected from three different datasets, and the results are given in Table 5 . The CNN with the best accuracy estimate is indicated in bold black.
Table 5.
Classification results of various proposed CNN (%).
| Acc | Sen | Spe | Prec | F_score | MCC | Kappa | Dice | Jaccard | Time (s) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Vgg16 | 98.94 | 98.21 | 99.22 | 98.79 | 98.50 | 97.76 | 97.62 | 98.32 | 96.70 | 142963.49 |
| Vgg19 | 98.42 | 91.23 | 98.70 | 97.98 | 97.60 | 96.52 | 96.44 | 97.59 | 95.29 | 84494.94 |
| Squeezenet | 97.23 | 95.06 | 97.83 | 96.96 | 95.98 | 94.06 | 93.76 | 95.70 | 91.75 | 17438.78 |
| Shufflenet | 97.82 | 96.67 | 98.41 | 97.40 | 97.03 | 95.52 | 95.10 | 96.56 | 93.35 | 24732.65 |
| Resnet101 | 97.99 | 96.43 | 98.43 | 97.63 | 97.02 | 95.63 | 95.47 | 96.89 | 93.96 | 112531.98 |
| Resnet50 | 97.95 | 96.75 | 98.36 | 97.53 | 97.14 | 95.68 | 95.40 | 96.71 | 93.62 | 69306.76 |
| Resnet18 | 98.09 | 97.36 | 98.71 | 97.22 | 97.29 | 95.95 | 95.69 | 96.96 | 94.10 | 26521.45 |
| GoogleNet | 97.52 | 97.00 | 98.45 | 96.35 | 96.67 | 94.96 | 94.43 | 96.06 | 92.42 | 29262.74 |
| DarkNet53 | 97.92 | 96.40 | 98.45 | 97.09 | 96.74 | 95.33 | 95.32 | 96.87 | 93.93 | 66582.46 |
| DarkNet19 | 98.68 | 97.75 | 98.99 | 98.60 | 98.16 | 97.24 | 97.03 | 97.89 | 95.86 | 58771.68 |
| AlexNet | 98.35 | 97.69 | 98.76 | 97.90 | 97.79 | 96.65 | 96.29 | 97.38 | 94.90 | 9617.79 |
| DenseNet201 | 97.76 | 96.40 | 98.33 | 97.16 | 96.77 | 95.21 | 94.95 | 96.50 | 93.23 | 92990.08 |
| InceptionResnet-V2 | 95.45 | 94.05 | 96.67 | 94.35 | 94.20 | 90.97 | 89.75 | 92.72 | 86.43 | 111230.22 |
Acc: Accuracy, Sen: Sensitivity, Spe: Specificity, Prec: Precision, MCC: Matthews correlation coefficient.
The confusion matrix of highest VGG16 and lowest InceptionResnet-V2 results are shown in Fig. 4 and Fig. 5 . Confusion matrix of the VGG16 network with dataset in Fig. 4, the Normal class achieves almost perfect classification precision, while the Viral Pneumonia class is classified a sensitivity of 93.31% in 269 test samples, classifying 1 sample as COVID-19 and 2 samples as Normal. In Normal class, it is the best sensitivity of 99.36% in 2038 test samples, and 4 samples are COVID-19 and 1 sample is Viral Pneumonia. In the COVID-19 class distribution, 4 samples are classified as Normal. Also, no instances are seen in the Viral Pneumonia class.
Fig. 4.
Confusion matrix of the VGG16 with the raw dataset.
Fig. 5.
Confusion matrix of the InceptionResnet-V2 with the raw dataset.
Table 6 shows the value of the detailed classification results of the best performing VGG16 for the raw dataset. The results are showed that MCC, precision, and F_score values of the COVID-19 class have values such as 98.00%, 98.34%, and 98.48%, respectively. The model with the raw dataset achieves 97.53%, 98.86%, and 97.74% accuracy values of MCC, precision, and F_score of Viral Pneumonia class, respectively. It indicates strong classification performance in Pneumonia and COVID-19, while usually the Normal class offers the highest accuracy.
Table 6.
Performance criteria of the trained Vgg16 in diagnosing the Chest X-rays (%).
| Sen | Spe | Prec | F_score | MCC | |
|---|---|---|---|---|---|
| COVID-19 | 98.62 | 99.48 | 98.34 | 98.48 | 98.00 |
| Normal | 99.36 | 98.29 | 99.17 | 99.26 | 97.75 |
| Viral Pneumonia | 96.65 | 99.89 | 98.86 | 97.74 | 97.53 |
The confusion matrix is shown with the raw dataset of InceptionResnet-V2 network in Fig. 5. Here, the Normal class achieves almost perfect classification precision, while the COVID-19 class has a sensitivity of 91.84% in 723 test samples, classifying 58 samples as Normal and 1 sample Viral Pneumonia. In Normal class, it is the best sensitivity of 99.36% in 2038 test samples, 44 samples are COVID-19 and 17 samples are Viral Pneumonia. The Viral Pneumonia class distribution is the best sensitivity of 93.31% in 269 test samples, 4 samples are COVID-19 and 14 samples are Normal.
In the predictive study, the InceptionResnet-V2 deep CNN belongs to the lowest performance and test results are interpreted in Table 7 . The prediction values of low COVID-19 are lower than the specificity, sensitivity, precision, F_score, and MCC performance criteria of the VGG16 deep learning network, which is the highest prediction performance value 1.56%, 7.07%, 5.16%, 6.03%, 7.92%, respectively. The prediction values of low Normal are lower than the sensitivity, specificity, precision, F_score, and MCC performance criteria of the VGG16 deep learning network, which is the highest prediction performance value, by 2.36%, 5.64%, 2.70%, 2.52%, 7.91%, respectively. The prediction values of low Viral Pneumonia are lower than the sensitivity, specificity, precision, F_score, and MCC performance criteria of the VGG16 deep learning network, which is the highest prediction performance value, by 3.45%, 0.54%, 5.61%, 4.53%, 5.00%, respectively. The predictive values in deep CNN are given in Table 7 range from VGG16 network to the values in InceptionResnet-V2 network.
Table 7.
Performance of the trained InceptionResnet-V2 in diagnosing the Chest X-rays.
| Sen | Spe | Prec | F_score | MCC | |
|---|---|---|---|---|---|
| COVID-19 | 91.84 | 97.92 | 93.26 | 92.54 | 90.23 |
| Normal | 97.01 | 92.74 | 96.49 | 96.75 | 90.01 |
| Viral Pneumonia | 93.31 | 99.35 | 93.31 | 93.31 | 92.66 |
The predictive results are analyzed using INCA and IRF feature selection to improve the results obtained in the predictive study using raw data in various deep CNN. As seen in Table 8 ; the prediction accuracy of the VGG16 network which has the best prediction value is 99.14% in the prediction study using INCA feature selection. Specificity and precision values are showed to improvement 99.56% and 98.80%, respectively. In the sensitivity value is observed 0.23% decrease. F_score, MCC, kappa, dice and Jaccard are observed to improve by 99.58%, 97.81%, 98.14%, 98.83% and 97.68%, respectively. The highest predictive accuracy on the Resnet18, Resnet50, and Resnet101 deep CNN has been 98.76% on the Resnet50 network, with a forecast time of approximately 6530.356s. According to the Resnet50 network value is calculated without feature selection, after the INCA feature selection is made, an increase of 0.82% is seen in the accuracy rate. The decrease in the prediction time that INCA feature selection in the Resnet50 network contributes to improving the results. Although there is an increase of 0.64% and 0.22% in sensitivity and Jaccard values, respectively, it is the lowest value compared to other performance criteria after feature selection. Although the DenseNet201 network an accuracy of 97.93%, improvement is observed in all performance criteria. Accuracy, specificity, precision, F_score, MCC, kappa, dice and Jaccard values are increased by 0.17%, 0.12%, 0.41%, 0.16%, 0.68%, 0.42%, 0.40% and 0.77%, respectively. The sensitivity value is decreased by 0.08%. GoogleNet, Shufflenet and VGG19 networks are observed to have 97.83%, 97.91% and 98.60% accuracy rates, respectively. Alexnet network is an accuracy of 98.86%, and it has a predictive time 9513.523s. Squzeenet network has an accuracy of 97.36%. Darknet19 and Darknet53 networks prediction accuracy is 99.02% and 98.26%, respectively. Darknet19 and Darknet53 networks accuracy rates increased by 0.34% in both algorithms. Although feature selection is applied, the InceptionResnet-V2 networks is showed a lower performance value of 96.01% compared to other algorithms. In addition, an increase in accuracy of 0.58% is observed. The lowest kappa and Jaccard performance criteria are showed 89.27% and 87.35%, respectively. It is predictive computational value is faster than other algorithms and has a value of 1005.125s.
Table 8.
Performance results of some deep CNN calculated with INCA feature selection (%).
| Acc | Sen | Spe | Prec | F_score | MCC | Kappa | Dice | Jaccard | Time (s) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Vgg16 | 99.14 | 97.98 | 99.56 | 98.80 | 99.58 | 97.8 | 98.1 | 98.83 | 97.6 | 12876.553 |
| Vgg19 | 98.60 | 91.10 | 98.88 | 98.16 | 97.75 | 97.21 | 96.85 | 98.32 | 96.69 | 7652.820 |
| Squeezenet | 97.36 | 95.86 | 98.23 | 97.54 | 96.12 | 94.58 | 94.23 | 95.75 | 93.47 | 1753.614 |
| Shufflenet | 97.91 | 96.95 | 98.62 | 98.56 | 97.40 | 95.93 | 95.85 | 96.72 | 93.64 | 2373.245 |
| Resnet101 | 98.12 | 97.35 | 99.21 | 98.16 | 97.23 | 96.21 | 95.82 | 96.95 | 94.08 | 11127.821 |
| Resnet50 | 98.76 | 97.38 | 98.75 | 97.98 | 97.56 | 95.79 | 95.96 | 96.82 | 93.83 | 6530.356 |
| Resnet18 | 98.54 | 97.77 | 99.12 | 97.86 | 97.75 | 96.15 | 95.96 | 97.12 | 94.40 | 2586.315 |
| GoogleNet | 97.83 | 96.85 | 98.75 | 96.74 | 96.81 | 95.18 | 94.73 | 96.58 | 93.38 | 2815.256 |
| DarkNet53 | 98.26 | 96.25 | 98.73 | 97.86 | 96.95 | 95.73 | 95.82 | 97.10 | 94.36 | 6585.325 |
| DarkNet19 | 99.02 | 97.12 | 99.17 | 98.85 | 98.56 | 97.87 | 97.25 | 98.23 | 96.52 | 5625.452 |
| AlexNet | 98.86 | 97.63 | 98.87 | 98.23 | 97.86 | 96.95 | 96.72 | 97.75 | 95.59 | 9513.523 |
| DenseNet201 | 97.93 | 96.32 | 98.45 | 97.56 | 96.93 | 95.87 | 95.36 | 96.89 | 93.96 | 9159.015 |
| InceptionResnet-V2 | 96.01 | 93.65 | 97.20 | 95.20 | 94.57 | 91.25 | 89.27 | 93.25 | 87.35 | 1005.125 |
As seen in Table 9 , IRF feature selection; The VGG16 network, which has the best predictive value, has a predictive result of 98.96% in the predictive study using IRF feature selection. Specificity and precision values are showed improvement of 98.85% and 99.01%, respectively. The sensitivity value is decreased by 0.47%. F_score, MCC, kappa, dice and Jaccard are improved, with 98.89%, 98.05%, 84.28%, 96.57% and 93.36%, respectively. The highest predictive accuracy on the Resnet18, Resnet50, and Resnet101 deep CNN is 98.30% on the Resnet18 network, with a forecast time of approximately 2572.103s. According to the Resnet18 value is calculated without feature selection, after the IRF feature selection is made, an increase of 0.21% is seen in the accuracy rate. This ratio is showed less accuracy than the Resnet50 accuracy estimate in INCA feature selection. The reduction of the prediction time of the Resnet18 network with IRF feature selection shows that the results of the analysis are improved. In addition, precision, F_score, MCC, kappa, dice and Jaccard performance metrics are calculated as 97.14%, 97.72%, 96.12%, 95.86%, 97.08% and 94.32%, respectively. We attained specificity value of 99.19% in the Resnet50 network. However, it has the highest predictive value of 11121.235s in the Resnet group. Although the DenseNet201 network has an accuracy of 97.86%, improvement is observed in all performance criteria. Accuracy, specificity, precision, F_score, MCC, kappa, dice and Jaccard values are increased by 0.10%, 0.03%, 0.37%, 0.12%, 0.53%, 0.35%, 0.25% and 0.50%, respectively. The sensitivity value is decreased by 0.005%. GoogleNet, Shufflenet, and VGG19 networks have an accuracy of 97.74%, 97.89%, and 98.06%, respectively. Alexnet network has an accuracy of 98.23%, and it is a predictive time 9514.632s. Squzeenet network has an accuracy of 97.30%. Darknet19 and Darknet53 networks prediction accuracy is 98.15% and 98.13%, respectively. Although feature selection is applied, the InceptionResnet-V2 network is showed a lower performance value of 95.60% compared to other algorithms. In addition, an increase in accuracy of 0.15% is observed. The lowest kappa and Jaccard performance criteria show 89.14% and 87.17%, respectively. It is predictive computational value which is faster than other algorithms and has a value of 1014.857s.
Table 9.
Performance results of some deep CNN calculated with IRF feature selection (%).
| Acc | Sen | Spe | Prec | F_score | MCC | Kappa | Dice | Jaccard | Time (s) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Vgg16 | 98.96 | 97.74 | 98.85 | 99.01 | 98.89 | 98.05 | 84.28 | 96.57 | 93.36 | 12885.12 |
| Vgg19 | 98.06 | 91.10 | 98.76 | 98.10 | 97.23 | 97.03 | 96.17 | 98.29 | 96.63 | 7663.270 |
| Squeezenet | 97.30 | 95.72 | 98.12 | 97.65 | 96.08 | 94.50 | 94.13 | 95.65 | 91.66 | 1745.853 |
| Shufflenet | 97.89 | 96.94 | 98.52 | 98.46 | 97.36 | 95.82 | 95.71 | 96.68 | 93.57 | 2371.265 |
| Resnet101 | 98.08 | 97.32 | 99.19 | 98.12 | 97.13 | 96.18 | 95.75 | 96.86 | 93.91 | 11121.23 |
| Resnet50 | 97.95 | 97.12 | 98.64 | 97.70 | 97.48 | 95.68 | 95.91 | 96.78 | 93.76 | 6528.20 |
| Resnet18 | 98.30 | 97.58 | 99.05 | 97.14 | 97.72 | 96.12 | 95.86 | 97.08 | 94.32 | 2572.10 |
| GoogleNet | 97.74 | 96.72 | 98.65 | 96.62 | 96.79 | 95.11 | 94.65 | 96.42 | 93.08 | 2802.10 |
| DarkNet53 | 98.13 | 96.17 | 98.65 | 97.75 | 96.90 | 95.69 | 95.78 | 97.06 | 94.28 | 6542.12 |
| DarkNet19 | 98.15 | 97.03 | 99.10 | 98.75 | 98.50 | 97.85 | 97.05 | 98.17 | 96.40 | 5623.25 |
| AlexNet | 98.23 | 97.61 | 98.56 | 98.12 | 97.80 | 96.92 | 96.68 | 97.71 | 95.52 | 9514.63 |
| DenseNet201 | 97.86 | 96.25 | 98.36 | 97.53 | 96.89 | 95.72 | 95.29 | 96.75 | 93.70 | 9160.00 |
| InceptionResnet-V2 | 95.60 | 93.23 | 97.12 | 95.12 | 94.48 | 91.14 | 89.14 | 93.15 | 87.17 | 1014.85 |
We selected a violin chart to present comparisons and display each quantitative metric in a single way by calculating the proposed segmentation quality measurement tools with the un-feature selection, INCA, and IRF methods over the Chest X-ray dataset. Violin charts show the probability of distribution of data at different values. The asymmetrical outer shape represents all possible outcomes. Also, as shown in Fig. 6 , the middle (+) toolbar shows the median value, and the green box shows the mean value of the data. Red, blue, and purple indicate segmentation results of un-feature selection, INCA and IRF, respectively. The comparison of the VGG16 network, which gave the best results from all analyzed deep CNN, with nine segmentation quality metrics is made. In Fig. 6, each of the nine separate graphs corresponds to three different distributions. Interestingly, the means between the three distributions and the intervals between the mean are different. Also, the distribution patterns are different. Accuracy (Fig. 6a), sensitivity (Fig. 6b), specificity (Fig. 6c), precision (Fig. 6d), F-score (Fig. 6e), MCC (Fig. 6f), kappa (Fig. 6g), dice (Fig. 6h) and Jaccard (Fig. 6ı) indicate a better segmentation performance. It is clear that the statistics of the accuracy, specificity, MCC, precision, and kappa metrics provided by INCA segmentation are greater than the un-feature selection and IRF methods. In addition, it is clear that the statistics of Dice, Jaccard, and sensitivity metrics provided by un-feature selection segmentation are larger than INCA and IRF methods.
Fig. 6.
Violin plots of the VGG16 network using accuracy, sensitivity, specificity, precision, F-score, MCC, kappa, dice, Jaccard, with suggested median values and mean against Un-feature selection, INCA and IRF segmentation methods.
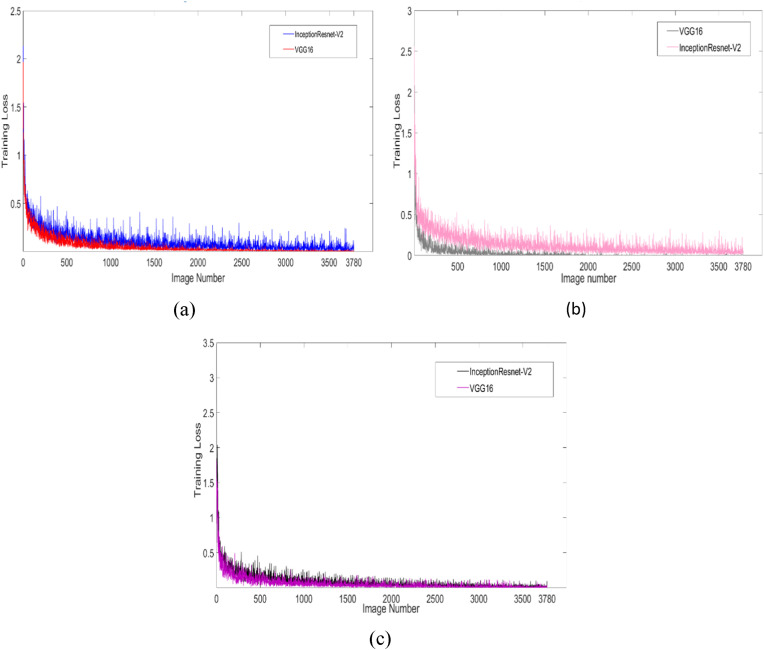
Prediction accuracy and performance criteria are calculated without any feature selection in the raw data in various deep CNN. Then the INCA and IRF feature selections are applied separately, and the graphs of the algorithm with the best results and the algorithm training losses with the worst results are shown in Fig. 7 . Fig. 7a shows the training loss in the calculation without any feature selection. This graph shows that the training loss of InceptionResnet-V2 network with low predictive accuracy is higher than VGG16 network with high training accuracy. After applying feature selection to the data, the highest estimation accuracy is observed in the VGG16 network, the lowest in the InceptionResnet-V2 network, the INCA and IRF feature selection training loss result graphs are shown in Fig. 7b and c.
Fig. 7.
Chart of non-feature selection (a), INCA feature selection (b) and IRF feature selection (c) deep CNN training loss.
4. Discussions
In this section, we compared between the pre-trained deep CNN proposed to detect COVID-19 and the models proposed in binary and multi-classification. Table 10 shows the highest data obtained in the studies mentioned. Seven different deep CNN and modified U-net network algorithms were used in the article [10], which consists of almost the same dataset in our study. In total, 18479 Chest X-ray images were used, and 3616 of these images are the same COVID-19 data. As a result of the analysis, sensitivity of 97.2%, accuracy of 96.29%, F1-score 96.28%. Modified U-net network, the accuracy of 98.63%, dice 96.94% predictive values were observed. Our proposed study is calculated the highest accuracy value as 98.94%, sensitivity 98.21%, and F1-score 98.50% in the VGG16 network without any feature selection. It is observed that VGG16 network performance criteria performed better after INCA and IRF feature selection. When compared with this study, it is seen that the INCA and IRF feature selection methods in our proposed study are improvement on performance criteria. In another study [44], COVID-19 diagnosis was studied in Inception-V3, Xception, and ResNet networks. In addition, 6432 Chest X-ray images were used in the study and 490 of these images were COVID-19 datasets. Compared to our study, less data was used as dataset. The highest values obtained are as seen in Table 10. Uçar and Korkmaz [11] performed a comprehensive analysis on a new Bayes-SqueezeNet network using a total of 5949 Chest X-ray images, 76 of which were COVID-19 data. As a result of the analysis, accuracy, F1_score and MCC performance criteria values were found 98.3%, 98.3% and 97.4%, respectively. In the study using the ResNet101 network [45], 5982 Chest X-ray images were used and 1765 of these images were COVID-19 data. The highest performance criteria were specificity, sensitivity and accuracy of 71.8%, 77.3% and 71.9%, respectively. In our proposed study; sensitivity, specificity and accuracy values of 15153 datasets analyzed without feature selection with the ResNet101 network is calculated as 96.43%, 98.43% and 97.99%, respectively. In addition, after the feature selection is applied in our proposed study, the performance criteria of the ResNet101 network are improved. In the study conducted using the ResNet34 network [25], the prediction accuracy value was calculated as 98.3%. 406 Chest X-ray images was analyzed, although the sensitivity value was better than the proposed study. In Ref. [46], COVID-19 detection of VGG19 and MobileNet-V2 networks were compared. In the analysis using the VGG19 network, the performance criteria of accuracy, recall and specificity were observed as 93.48%, 92.85% and 98.75%, respectively.
Table 10.
Comparison of the lung Chest X-ray models for multiclass classification.
| Ref. | Method | Dataset |
Performance (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Covid | Total | Acc | Sen | Spe | Prec | F1_ score | Dice | MCC | Kappa | Jaccard | ||
| [10] | Seven different CNN networks | 3616 | 18479 | 96.29 | 97.2 | – | 96.28 | |||||
| Unet network | 3616 | 18479 | 98.63 | 96.94 | ||||||||
| [44] | Inception-V3, Xception, ResNet |
490 | 6432 | 96.00 | 92.00 | |||||||
| [11] | Bayes-SqueezeNet | 76 | 5949 | 98.3 | 98.3 | 97.4 | ||||||
| [45] | ResNet101 | 1765 | 5982 | 71.9 | 77.3 | 71.8 | – | – | – | – | – | – |
| [25] | ResNet34 | 203 | 406 | 98.3 | 1 | 96.67 | 96.7 | 98.36 | – | – | – | – |
| [46] | VGG19 | 224 | 1427 | 93.48 | 92.85 | 98.75 | ||||||
| [46] | MobileNet v2 | 224 | 1427 | 92.85 | 99.10 | 97.09 | ||||||
| Our study | Thirteen different CNN networks | 3616 | 15153 | 99.14 | 97.98 | 99.56 | 98.8 | 99.58 | 98.83 | 97.8 | 98.1 | 97.6 |
5. Conclusion
This study presents a deep CNN approach for the automatic detection of COVID-19 pneumonia. The dataset is consisted of a total of 15153 Chest X-ray images. Thirteen different popular and previously reported effective deep CNN are trained and tested to classify normal, COVID-19 and viral pneumonia patients using Chest X-ray images. The features obtained from these deep CNN are given as input data to the INCA and IRF feature selection algorithms. The features obtained after the analysis of thirteen different deep CNN with INCA and IRF feature selection methods were given to the deep CNN as input data again. The specified performance criteria are calculated. Although the VGG16 network shows the best performance criterion with a value of 98.94%, the worst performance criterion is InceptionResnet-V2 network with a value of 95.45%. First of all, the features extracted from the analysis are classified. Then, these features are reclassified using INCA and IRF feature selection algorithms. Deep CNN with the best and worst results hasn't changed after INCA feature selection. After the INCA feature selection, the accuracy value of the VGG16 network is increased to 99.14%. The accuracy of the InceptionResnet-V2 network is increased by 96.01%. Deep CNN with the best and worst results hasn't changed after IRF feature selection. After the INCA feature selection, the accuracy value of the VGG16 network is increased to 98.96%. The accuracy value of the InceptionResnet-V2 network is increased by 95.60%. Accuracy, specificity, sensitivity, precision, F-score, MCC, kappa, dice, Jaccard, with suggested median values and mean against Un-feature selection, using INCA and IRF segmentation methods are presented violin plots. However, training losses are observed during Un-feature selection, INCA and IRF segmentation. When the data are analyzed without feature selection, the highest accuracy result is 98.94% in VGG16 network. Sensitivity, specificity, precision, F_score and Dice values are 98.21%, 99.22%, 98.79%, 98.50% and 98.32% respectively. MCC, Kappa and Jaccard performance criteria values are 97.76%, 97.62% and 96.70%, respectively. In the analysis made with the INCA feature selection method, the VGG16 network is the best accuracy of 99.14% performance criteria. Specificity, precision, F_score, Kappa and Dice values are 99.56%, 98.80%, 99.56%, 98.14% and 98.83% respectively. Sensitivity, MCC, Jaccard performance criteria values are 97.98%, 97.81% and 97.68%, respectively. In the analysis made with the IRF feature selection method, the VGG16 network has the best accuracy of 98.96% performance criteria. Specificity, precision, F_score and MCC performance criteria values are 98.85%, 99.01%, 98.89% and 98.05%, respectively. Sensitivity, Kappa, Dice, Jaccard performance criteria values are 97.74%, 84.28%, 96.57% and 93.36%, respectively.
In future, we intend to refine our techniques and suggest new techniques as more real data become available. In addition, different feature selection or feature extraction methods can be developed and tested on a dataset with different features.
Funding
The authors state that this work has not received any funding.
CRediT authorship contribution statement
Narin Aslan: Visualization, Writing – review & editing, Writing – original draft, Data curation, Resources, Investigation, Formal analysis, Validation, Software, Conceptualization, Methodology. Gonca Ozmen Koca: Project administration, Supervision, Visualization, Writing – review & editing, Writing – original draft, Data curation, Resources, Formal analysis, Validation, Methodology, Conceptualization. Mehmet Ali Kobat: Writing – original draft, Validation, Formal analysis, Conceptualization, Writing – review & editing. Sengul Dogan: Project administration, Supervision, Writing – review & editing, Writing – original draft, Formal analysis, Validation, Methodology, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.World Health Organization . Emergencies Preparedness, Response, Disease Outbreak News. World Health Organization (WHO); 2020. Pneumonia of unknown cause–China. [Google Scholar]
- 2.Li Y., Yao L., Li Jiawei, Chen Lei, et al. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J. Med. Virol. 2020:1–6. doi: 10.1002/jmv.25786. Wiley. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y., Kang H., Liu X., Tong Z. Combination of RT-PCR testing and clinical features for diagnosis of COVID-19 facilitates management of SARSCoV-2 outbreak. J. Med. Virol. 2020;22(6):538–539. doi: 10.1002/jmv.25721. (Wiley) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zebin T., Rezvy S. COVID-19 detection and disease progression visualization: deep learning on chest X-rays for classification and coarse localization. Appl. Intell. 2021;51(2):1010–1021. doi: 10.1007/s10489-020-01867-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karar M.E., Hemdan E.E.D., Shouman M.A. Cascaded deep learning classifiers for computer-aided diagnosis of COVID-19 and pneumonia diseases in X-ray scans. Complex & Intelligent Systems. 2021;7(1):235–247. doi: 10.1007/s40747-020-00199-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maghdid H.S., Asaad A.T., Ghafoor K.Z., Sadiq A.S., Mirjalili S., Khan M.K. vol. 11734. International Society for Optics and Photonics; 2021. April). Diagnosing COVID-19 pneumonia from X-ray and CT images using deep learning and transfer learning algorithms; p. 117340E. (Multimodal Image Exploitation and Learning). 2021. [Google Scholar]
- 7.Khan M.A., Kadry S., Zhang Y.D., Akram T., Sharif M., Rehman A., Saba T. Prediction of COVID-19-pneumonia based on selected deep features and one class kernel extreme learning machine. Comput. Electr. Eng. 2021;90:106960. doi: 10.1016/j.compeleceng.2020.106960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tarik A., Aissa H., Yousef F. Artificial intelligence and machine learning to predict student performance during the COVID-19. Procedia Comput. Sci. 2021;184:835–840. doi: 10.1016/j.procs.2021.03.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 10.Rahman T., Khandakar A., Qiblawey Y., Tahir A., Kiranyaz S., Kashem S.B.A.…Chowdhury M.E. Exploring the effect of image enhancement techniques on COVID-19 detection using chest X-ray images. Comput. Biol. Med. 2021;132:104319. doi: 10.1016/j.compbiomed.2021.104319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ucar F., Korkmaz D. COVIDiagnosis-Net: deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med. Hypotheses. 2020;140:109761. doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahmud T., Rahman M.A., Fattah S.A. CovXNet: a multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput. Biol. Med. 2020;122:103869. doi: 10.1016/j.compbiomed.2020.103869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nour M., Cömert Z., Polat K. A novel medical diagnosis model for COVID-19 infection detection based on deep features and Bayesian optimization. Appl. Soft Comput. 2020;97:106580. doi: 10.1016/j.asoc.2020.106580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tuncer T., Ozyurt F., Dogan S., Subasi A. A novel Covid-19 and pneumonia classification method based on F-transform. Chemometr. Intell. Lab. Syst. 2021;210:104256. doi: 10.1016/j.chemolab.2021.104256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020;121:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ozyurt F., Tuncer T., Subasi A. An automated COVID-19 detection based on fused dynamic exemplar pyramid feature extraction and hybrid feature selection using deep learning. Comput. Biol. Med. 2021;132:104356. doi: 10.1016/j.compbiomed.2021.104356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marques G., Agarwal D., de la Torre Díez I. Automated medical diagnosis of COVID-19 through EfficientNet convolutional neural network. Appl. Soft Comput. 2020;96:106691. doi: 10.1016/j.asoc.2020.106691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abbas A., Abdelsamea M.M., Gaber M.M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl. Intell. 2021;51(2):854–864. doi: 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karakanis S., Leontidis G. Lightweight deep learning models for detecting COVID-19 from chest X-ray images. Comput. Biol. Med. 2021;130:104181. doi: 10.1016/j.compbiomed.2020.104181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ismael A.M., Şengür A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst. Appl. 2021;164:114054. doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zebin T., Rezvy S. COVID-19 detection and disease progression visualization: deep learning on chest X-rays for classification and coarse localization. Appl. Intell. 2021;51(2):1010–1021. doi: 10.1007/s10489-020-01867-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Falluji R.A., Katheeth Z.D., Alathari B. Automatic detection of COVID-19 using chest X-ray images and modified ResNet18-based convolution neural networks. Comput. Mater. Continua (CMC) 2021:1301–1313. [Google Scholar]
- 23.Karar M.E., Hemdan E.E.D., Shouman M.A. Cascaded deep learning classifiers for computer-aided diagnosis of COVID-19 and pneumonia diseases in X-ray scans. Complex & Intelligent Systems. 2021;7(1):235–247. doi: 10.1007/s40747-020-00199-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saha P., Sadi M.S., Islam M.M. EMCNet: automated COVID-19 diagnosis from X-ray images using convolutional neural network and ensemble of machine learning classifiers. Informatics in medicine unlocked. 2021;22:100505. doi: 10.1016/j.imu.2020.100505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nayak S.R., Nayak D.R., Sinha U., Arora V., Pachori R.B. Application of deep learning techniques for detection of COVID-19 cases using chest X-ray images: a comprehensive study. Biomed. Signal Process Control. 2021;64:102365. doi: 10.1016/j.bspc.2020.102365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Demir F. DeepCoroNet: a deep LSTM approach for automated detection of COVID-19 cases from chest X-ray images. Appl. Soft Comput. 2021;103:107160. doi: 10.1016/j.asoc.2021.107160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saygılı A. A new approach for computer-aided detection of coronavirus (COVID-19) from CT and X-ray images using machine learning methods. Appl. Soft Comput. 2021;105:107323. doi: 10.1016/j.asoc.2021.107323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ibrahim D.M., Elshennawy N.M., Sarhan A.M. Deep-chest: multi-classification deep learning model for diagnosing COVID-19, pneumonia, and lung cancer chest diseases. Comput. Biol. Med. 2021;132:104348. doi: 10.1016/j.compbiomed.2021.104348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maghdid H.S., Asaad A.T., Ghafoor K.Z., Sadiq A.S., Mirjalili S., Khan M.K. vol. 11734. International Society for Optics and Photonics; 2021. April). Diagnosing COVID-19 pneumonia from X-ray and CT images using deep learning and transfer learning algorithms; p. 117340E. (Multimodal Image Exploitation and Learning). 2021. [Google Scholar]
- 30.Toğaçar M., Ergen B., Cömert Z. Detection of lung cancer on chest CT images using minimum redundancy maximum relevance feature selection method with convolutional neural networks. Biocybernetics and Biomedical Engineering. 2020;40(1):23–39. [Google Scholar]
- 31.Amyar A., Modzelewski R., Li H., Ruan S. Multi-task deep learning based CT imaging analysis for COVID-19 pneumonia: classification and segmentation. Comput. Biol. Med. 2020;126:104037. doi: 10.1016/j.compbiomed.2020.104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loey M., Manogaran G., Khalifa N.E.M. A deep transfer learning model with classical data augmentation and cgan to detect covid-19 from chest ct radiography digital images. Neural Comput. Appl. 2020:1–13. doi: 10.1007/s00521-020-05437-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Budak U., Cibuk M., Comert Z., Sengur A. Efficient COVID-19 segmentation from CT slices exploiting semantic segmentation with integrated attention mechanism. J. Digit. Imag. 2021:1–10. doi: 10.1007/s10278-021-00434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahuja S., Panigrahi B.K., Dey N., Rajinikanth V., Gandhi T.K. Deep transfer learning-based automated detection of COVID-19 from lung CT scan slices. Appl. Intell. 2021;51(1):571–585. doi: 10.1007/s10489-020-01826-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.https://sirm.org/category/senza-categoria/covid-19,(Access date:06/03/2021).
- 36.https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia, (Access date:06/03/2021).
- 37.https://www.kaggle.com/c/rsna-pneumonia-detection-challenge/data,(Access date:06/03/2021).
- 38.Raghu S., Sriraam N. Classification of focal and non-focal EEG signals using neighborhood component analysis and machine learning algorithms. Expert Syst. Appl. 2018;113:18–32. [Google Scholar]
- 39.Jin M., Deng W. Predication of different stages of Alzheimer's disease using neighborhood component analysis and ensemble decision tree. J. Neurosci. Methods. 2018;302:35–41. doi: 10.1016/j.jneumeth.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 40.Tuncer T., Dogan S., Özyurt F., Belhaouari S.B., Bensmail H. Novel multi center and threshold ternary pattern based method for disease detection method using voice. IEEE Access. 2020;8:84532–84540. [Google Scholar]
- 41.Tuncer T., Dogan S., Acharya U.R. Automated accurate speech emotion recognition system using twine shuffle pattern and iterative neighborhood component analysis techniques. Knowl. Base Syst. 2021;211:106547. [Google Scholar]
- 42.Karabulut N., Ozmen Koca G. Wave height prediction with single input parameter by using regression methods. Energy Sources, Part A Recovery, Util. Environ. Eff. 2020;42(24):2972–2989. [Google Scholar]
- 43.Tuncer T., Dogan S., Ozyurt F. An automated residual exemplar local binary pattern and iterative ReliefF based COVID-19 detection method using chest X-ray image. Chemometr. Intell. Lab. Syst. 2020;203:104054. doi: 10.1016/j.chemolab.2020.104054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jain R., Gupta M., Taneja S., Hemanth D.J. Deep learning based detection and analysis of COVID-19 on chest X-ray images. Appl. Intell. 2021;51(3):1690–1700. doi: 10.1007/s10489-020-01902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Che Azemin M.Z., Hassan R., Mohd Tamrin M.I., Md Ali M.A. COVID-19 deep learning prediction model using publicly available radiologist-adjudicated chest X-ray images as training data: preliminary findings. Int. J. Biomed. Imag. 2020 doi: 10.1155/2020/8828855. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Physical and Engineering Sciences in Medicine. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]