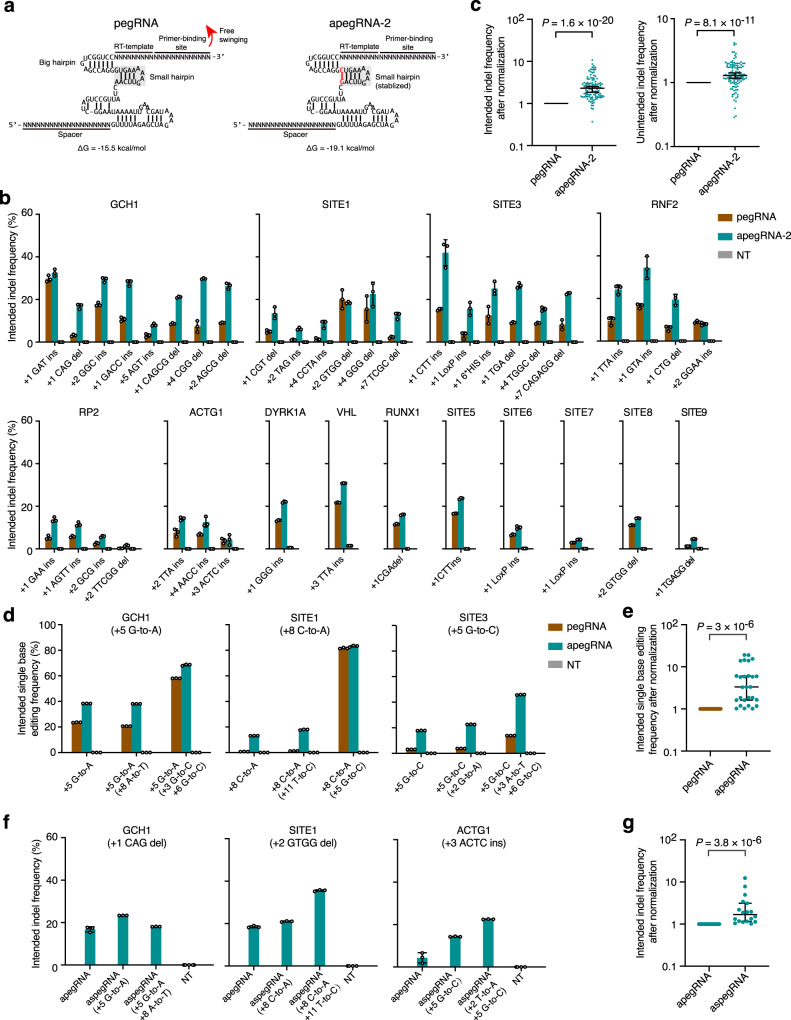
Fig. 2. pegRNA with a stabilized secondary structure induced higher efficiencies of intended indel and single-base editing.
a Schematic diagrams illustrating the predicted secondary structures of regular pegRNA and apegRNA-2. Presumably, the free swinging of the RTT and PBS can break up the small hairpin (left panel), which destabilizes pegRNA. However, engineering within the small hairpin of pegRNAs can stabilize the secondary structures of apegRNA-2 (right panel). b Intended indel frequencies were induced by pegRNA and apegRNA-2 at the indicated target sites under the PE3 setting or from non-transfected (NT) cells. c Statistical analyses of the intended indel frequencies and unintended indel frequencies after normalization, setting the frequencies induced by regular pegRNAs as 1. n = 117 editing from three independent experiments shown in b. d Intended single-base editing frequencies were induced by pegRNA and apegRNA without or with additional base substitutions at the indicated target sites under the PE3 setting. e Statistical analysis of normalized single base editing frequencies, setting the frequencies induced by regular pegRNAs as 1. n = 18 editing from three independent experiments shown in d. f Intended indel frequencies were induced by apegRNA and aspegRNA (with additional base substitutions) at the indicated target sites under the PE3 setting. g Statistical analysis of normalized intended indel frequencies, setting the frequencies induced by apegRNA without additional base substitution as 1. n = 18 editing from three independent experiments shown in f. b, d, f Means ± s.d. are from three independent experiments. c, e, g P value, Wilcoxon one-tailed signed-rank test. The median and interquartile range (IQR) are shown. Source data are provided as a Source Data file.