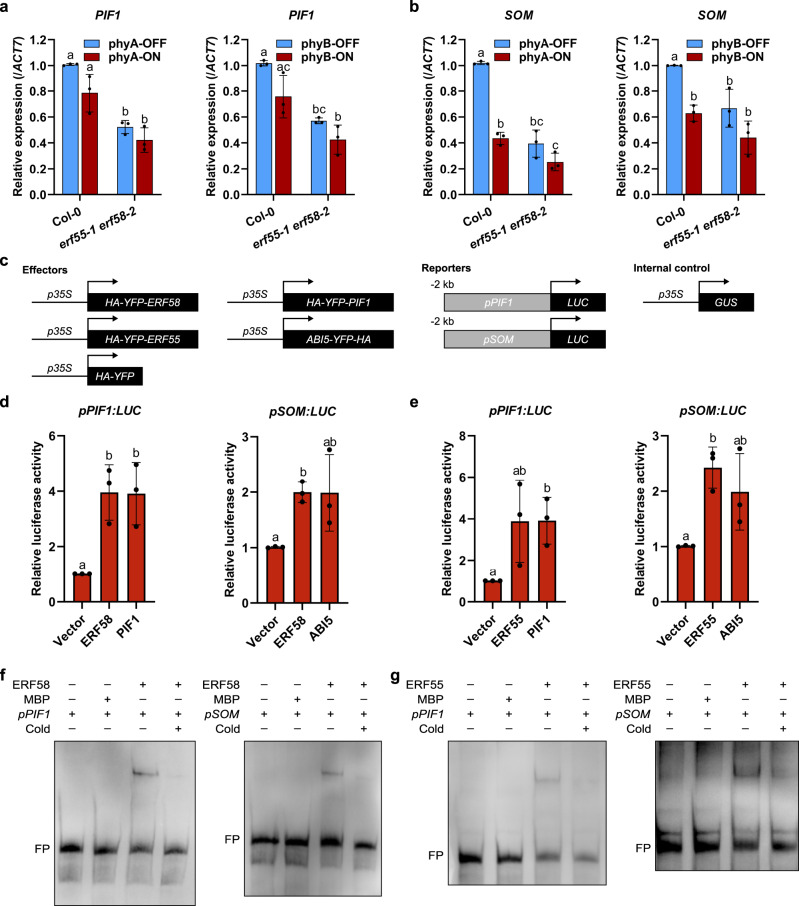
Fig. 3. ERF58 directly binds to the promoter of PIF1 and SOM to induce their expression.
a, b RT-qPCR analysis of PIF1 (a) and SOM (b) expression in Col-0 and erf55-1 erf58-2 seeds germinating under phyA-ON/OFF or phyB-ON/OFF conditions. Light conditions are described in Supplementary Fig. 6a, b. ACT7 was used as an internal control. Values are means of three replicates ±SD. Different letters indicate significant differences as determined by two-way ANOVA followed by post-hoc Tukey’s HSD test; p < 0.05. c Constructs used for transactivation assays. d, e Transactivation assay in tobacco. Combinations of effector (p35S:HA-YFP-ERF58 or p35S:HA-YFP-ERF55; p35S:HA-YFP was used as negative control; p35S:HA-YFP-PIF1 and p35S:ABI5-YFP-HA were used as positive controls) and reporter constructs (pPIF1 −1…−2000:LUC or pSOM −1…−2000:LUC) and p35S:GUS (used for normalisation) were transiently co-expressed in tobacco leaf epidermis cells by agro infiltration. LUC and GUS activity was measured in protein extracts from infiltrated leaves. Bars show mean relative LUC activity (LUC activity divided by GUS activity) of three replicates ±SD. Different letters indicate significant differences as determined by t-test and Holm–Bonferroni method; p < 0.05. f, g EMSAs. Biotin-labelled PIF1 and SOM promoter fragments (pPIF1 −801…−1031 and pSOM −621…−840) were incubated with MBP-ERF58, MBP-ERF55, or MBP alone (negative control). Samples were analysed by native PAGE. Gels were blotted onto nylon membranes and signals were detected by streptavidin-coupled horseradish peroxidase and ECL. FP, free probe; Cold, excess of unlabelled DNA fragment containing the DRE motif to which ERF55/ERF58 bind. Experiments were repeated at least three times with similar results.