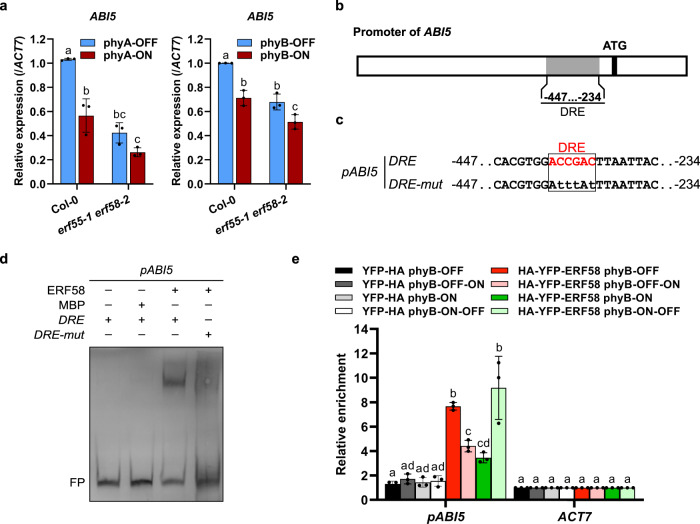
Fig. 6. ERF55 and ERF58 upregulate ABI5 expression.
a RT-qPCR analysis of ABI5 expression levels in Col-0 and erf55-1 erf58-2 seeds germinating under phyA-ON/OFF or phyB-ON/OFF conditions. Light conditions are described in Supplementary Fig. 6a, b. ACT7 was used as an internal control. Values show means of three replicates ±SD. b, c Schematic representation of the ABI5 promoter. The promoter fragment used for EMSA is shown in c. d EMSA. Biotin-labelled ABI5 −234…−447 promoter fragments containing a wild-type or mutated DRE motif were incubated with MBP-ERF58 or MBP alone (negative control). Samples were analysed by native PAGE. The gel was blotted onto a nylon membrane and signals were detected by streptavidin-coupled horseradish peroxidase and ECL. FP, free probe. The experiment was repeated five times with similar results. Data for ERF55 are shown in Supplementary Fig. 13b. e ChIP-qPCR. Germinating seeds of Col-0 p35S:HA-YFP-ERF58 (line 21C) and Col-0 p35S:YFP-HA (negative control) were subject to light treatments as described in Fig. 4a. Chromatin was isolated and HA-YFP-ERF58 bound DNA fragments were purified using α-GFP coupled magnetic beads. qPCR and specific primers were used to detect the ABI5 −234…−447 promoter fragment in the eluate fraction. ACT7 was used for normalisation. Values show enrichment of the ABI5 promoter fragment in the eluate fraction compared to the input fraction and are means of three replicates ±SD. a, e Different letters indicate significant differences as determined by two-way ANOVA followed by post-hoc Tukey’s HSD test. a p < 0.05. e p < 0.01.