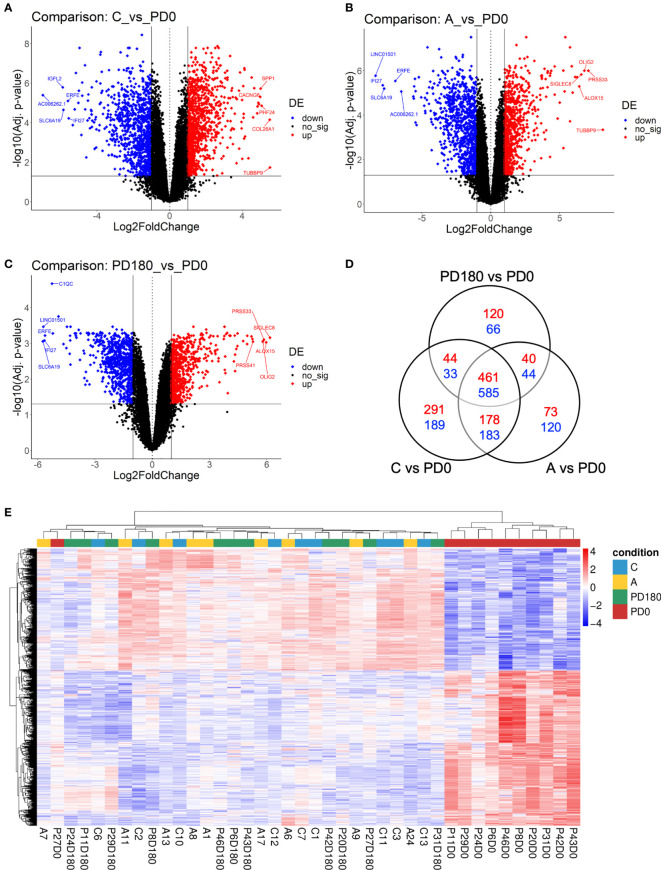
Figure 3.
Differential expression analysis comparing VL patients to nondiseased patients identified hundreds of differentially expressed genes (DEGs). Volcano plots highlighting genes significantly regulated (FDR <0.05, horizontal threshold line set on y-axis) in the comparisons C vs. PD0 (A), A vs. PD0 (B) and PD0 vs. PD180 (C). Vertical lines at -1 and +1 on the x-axis indicate the expression level criteria of fold decrease or increase, respectively, applied to DEGs that were further analyzed (all genes colored in blue or red); (D) Venn diagram displaying the number of exclusive DEGs for each comparison, as well as the number of shared DEGs among them. Dashed squares indicate the DEG lists used as subsets of prioritized genes in the mRNA-lncRNA coexpression profile analysis; (E) Heatmap of 1,045 DEGs shared among the three comparisons (suggested to be the gene signature of VL disease status), depicting the clustering of samples in two major groups: VL patients (PD0 labels, in red), the very consistent cluster on the right side and a heterogeneous cluster encompassing the nondiseased groups (C, A and PD180 labels, in blue, yellow and green, respectively) split into minor clusters. Z scores of cpm read counts were used, and a graded color scale from red to blue indicates whether the level of gene expression was above or below the mean (i.e., up- or downregulation).