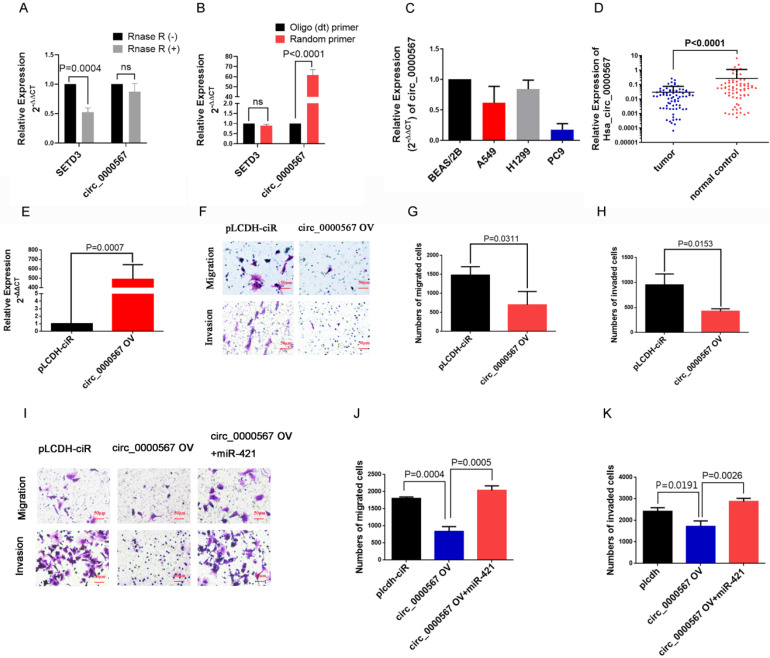
Figure 5.
circ_0000567 suppresses migration and invasion of miR-421 by sponging miR-421. A circ_0000567 was resistant to RNase R, whereas the linear SETD3 transcript could be degraded by RNase R. B circ_0000567 could only be reverse transcribed using random primers, but not oligo(dT) primers. However, there was no difference between using random primers or oligo(dT) primers in reverse transcription of the linear SETD3 transcript. C circ_0000567 was downregulated in lung adenocarcinoma (LUAD) cell lines, including A549, PC9, and H1299, compared to control BEAS/2B cells. D circ_0000567 was verified to be downregulated in the 73 LUAD specimens compared to the paired normal lung tissues (P<0.0001). E qRT-PCR analysis of circ_0000567 expression after treatment with circ_0000567 overexpression plasmids. F-H circ_0000567 reduced cell migration and invasion as determined by transwell assays with and without Matrigel. I-K miR-421 partially abolished the effects of circ_0000567 on cell migration and invasion, as revealed by transwell assay in LUAD cells.