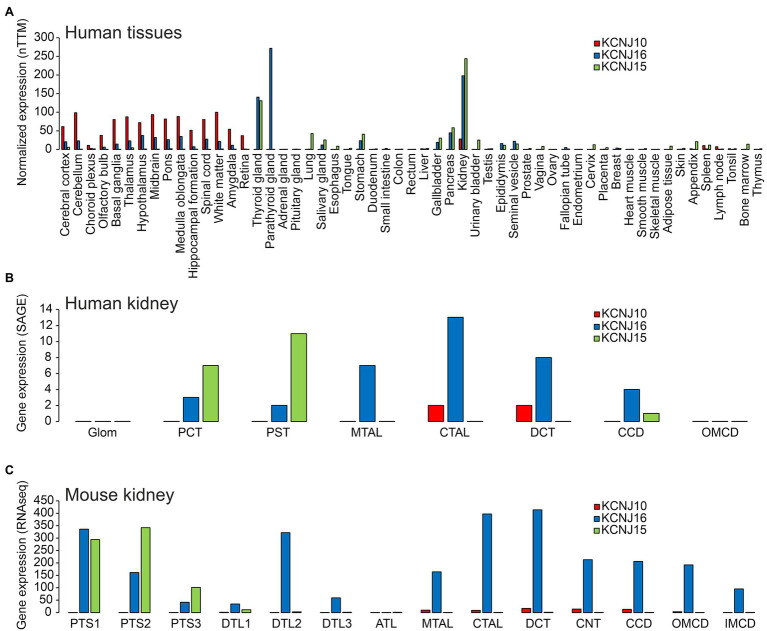
Figure 2.
(A) Expression of KCNJ10 (Kir4.1), KCNJ16 (Kir5.1), and KCNJ15 (Kir4.2) in human tissues. Data were obtained from “The Human Protein Atlas” (https://www.proteinatlas.org/ENSG00000177807-KCNJ10/summary/rna; https://www.proteinatlas.org/ENSG00000153822-KCNJ16/summary/rna; https://www.proteinatlas.org/ENSG00000157551-KCNJ15/summary/rna). Data are presented as “consensus dataset” consisting of normalized expression (nTPM) levels, created by combining the HPA and GTEx transcriptomics datasets using the internal normalization pipeline of “The Human Protein Atlas” (https://www.proteinatlas.org). Please note that the number of samples for each tissue and the origin of the different tissues is different. Therefore, it can be difficult to quantitatively compare expression in different tissues. (B) Expression of KCNJ10, KCNJ16, and KCNJ15 along the human nephron. Data are taken from (Chabardes-Garonne et al., 2003). In this beautiful study, fresh human kidney tissue from nine donors was microdissected into glomeruli (glom), proximal convoluted tubules (PCT), proximal straight tubules (PST), medullary thick ascending limbs (MTAL), cortical thick ascending limbs (CTAL), distal convoluted tubules (DCT), cortical collecting ducts (CCD), and outer medullary collecting ducts (OMCD). Gene expression was assessed by “serial analysis of gene expression” (SAGE). (C) Expression of KCNJ10, KCNJ16, and KCNJ15 along the mouse tubular system. Data are taken from https://esbl.nhlbi.nih.gov/MRECA/Nephron/ (Chen et al., 2021). In this beautiful study, RNA-seq was performed on murine microdissected tubular segments. Initial segment of the proximal tubule (PTS1); proximal straight tubule in cortical medullary rays (PTS2); last segment of the proximal straight tubule in the outer stripe of outer medulla (PTS3); short descending limb of the loop of Henle (DTL1); long descending limb of the loop of Henle in the outer medulla (DTL2); long descending limb of the loop of Henle in the inner medulla (DTL3); thin ascending limb of the loop of Henle (ATL); medullary thick ascending limb of the loop of Henle (MTAL); cortical thick ascending limb of the loop of Henle (CTAL); macula densa (MD); distal convoluted tubule (DCT); connecting tubule (CNT); cortical collecting duct (CCD); outer medullary collecting duct (OMCD); inner medullary collecting duct (IMCD). Another excellent source of transcriptome data is https://cello.shinyapps.io/kidneycellexplorer/, which provides single cell RNA-seq data of mouse kidney (Ransick et al., 2019). These single cell-based data indicate that Kir4.1 and Kir5.1 are strongly expressed in principal cells of connecting tubules and collecting ducts, but not in intercalated cells (data not shown).