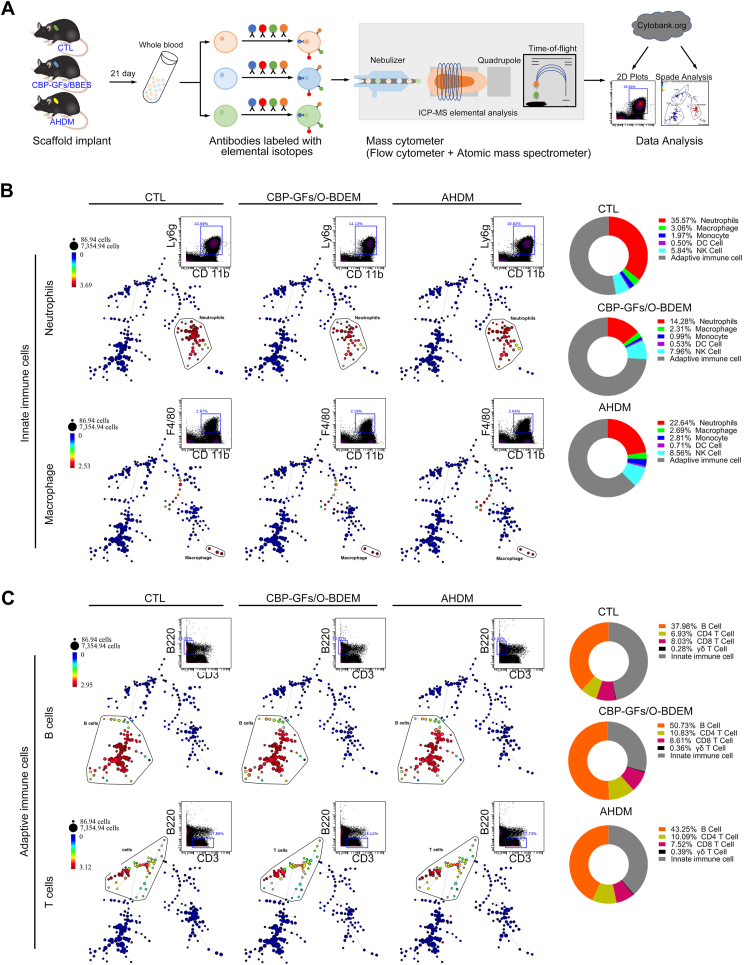
Fig. 6.
Differential abundances of systematic innated and adaptive immune cell populations in the mice blood samples from CTL, CBP-GFs/O-BDEM and AHDM groups. (A) Schematic diagram of the experimental procedure. Mice blood samples from CTL, CBP-GFs/O-BDEM and AHDM groups were collected respectively at postoperative 21-day. After lysis of the red blood cells, these pooled cells were stained with antibodies conjugated to elemental isotopes, and then analyzed using a CyTOF machine. The immune cell populations were manually gated, and then identified using SPADE algorithms. (B) The SPADE tree and dot plots analysis shows the abundance of CD11b+Ly6g+ neutrophils or F4/80+ macrophage in each group. In the SPADE diagram, the number of immune cell populations were represented with node size, and the colored gradient of the node correlates with the median intensity of marker-expression. The relative abundance of innate immune cells (monocytes, macrophages, neutrophils, DC cells and NK cells) among three groups, as calculated from the SPADE analysis. (C) The SPADE tree and dot plots analysis shows the frequency of B and T cells in CTL, CBP-GFs/O-BDEM and AHDM groups. The relative abundance of adaptive immune cells (B cells, CD4+T cells, CD8+T cells, and γδT cells) among three group calculated from the SPADE analysis. All data are presented as the mean ± SEM (n = 3).