Figure 5. HIV-1 NC ZnF Mutants Reverse the HIV-1 SG-Blockade and Induce vRNA Localization to SGs.
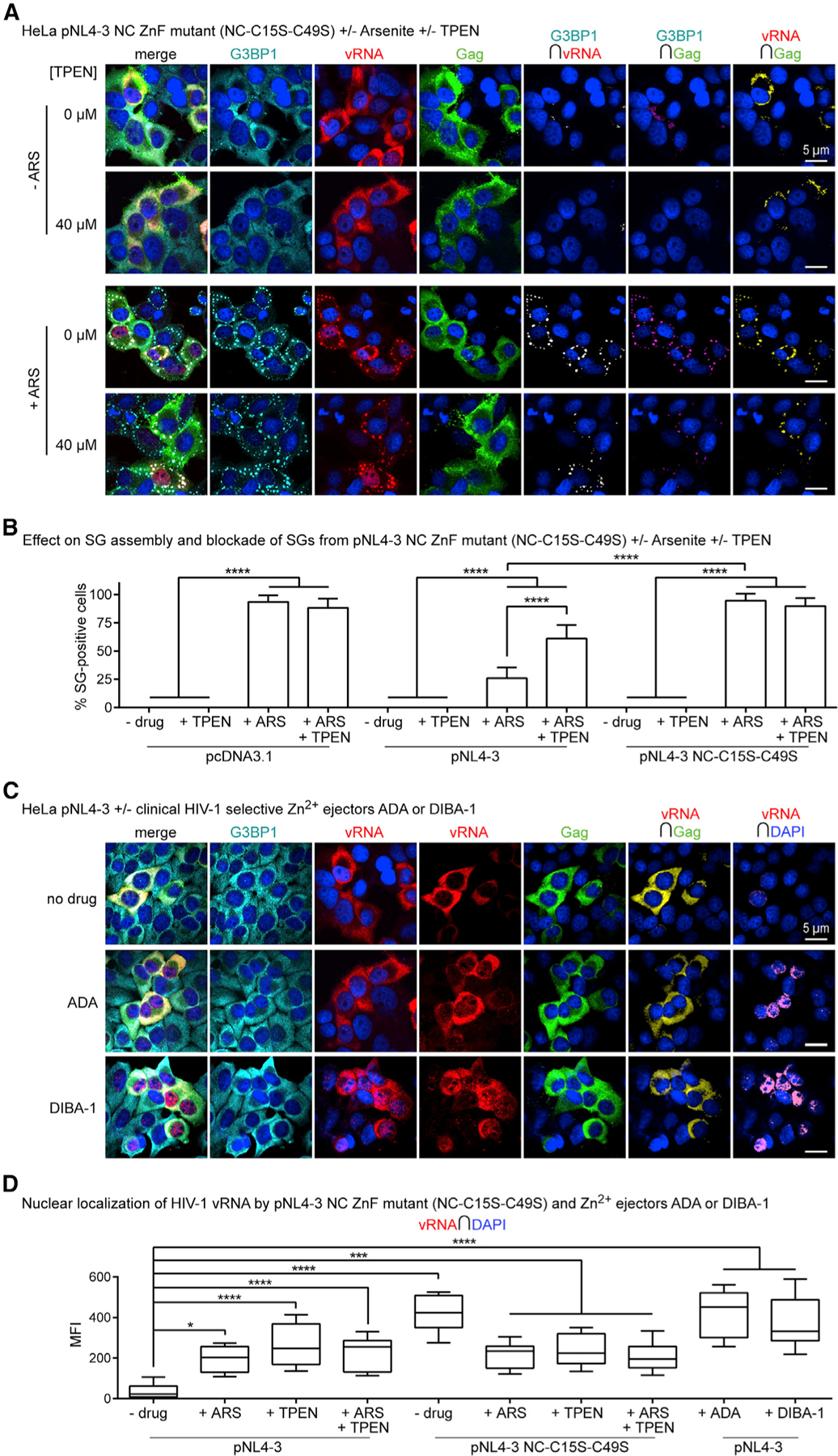
(A) HeLa cells transfected with pNL4–3 NC ZnF mutant (NC-C15S-C39S) and treated with ARS (500 µM) and/or TPEN (40 µM) for effects on SG assembly and localization of G3BP1 (cyan), vRNA (red), and Gag (green), where NC ZnF mutant cannot block SG assembly and restricts the vRNA to SGs and nuclei (n = 4).
(B) Bar graph comparing SG numbers in HeLa cells across conditions tested, including data presented in Figure 3B; demonstrating that the pNL4–3 NC ZnF mutant cannot block SG assembly relative to WT pNL4–3 transfected cells, which is unaltered by TPEN (40 µM) (n = 4).
(C) HeLa cells transfected with WT pNL4–3 and treated with ADA (100 µM) or DIBA-1 (50 µM) for their effects on localization of G3BP1 (cyan), vRNA (red), and Gag (green), where both drugs cause the nuclear retention of the vRNA (n = 3).
(D) Boxplots of vRNA-DAPI colocalization comparing effects of ADA or DIBA-1 with other conditions tested; demonstrating that all Zn2+ drugs cause nuclear retention of the vRNA (−drug versus +ADA; −drug versus DIBA; −drug versus TPEN; pNL4–3 versus pNL4–3 NC-C15S-C49S) (n = 3). Statistical analysis details described in STAR Methods. Boxplot horizontal lines indicate median, and whiskers are minimum to maximum; ∩, colocalization (intersection); MFI, mean fluorescence intensity.
