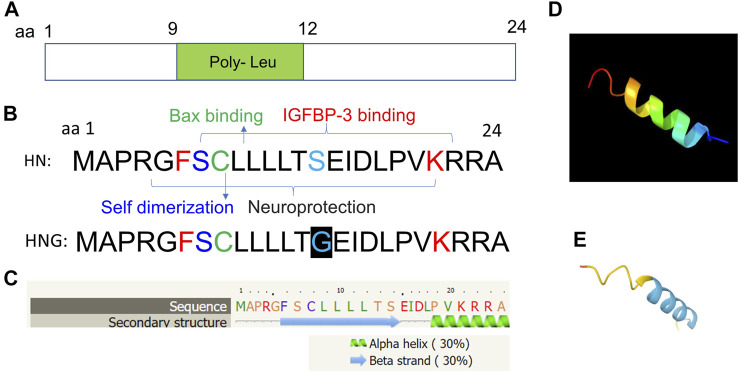
FIGURE 3.
Predicted molecular structural analyses of HN. (A) HN is a small peptide of 24 amino acid residue and is comprised of a poly-leu motif (amino acid residues 9–12). (B) Key residues for BAX binding (amino acid residue 8), IGFBP3 binding (amino acid residues 6–21), and self-dimerization (amino acid residue 7) are shown. The full sequence of HN and its analogue HNG (S14G) with substitution of serine at amino acid residue 14 to glycine are shown. (C) Secondary structure predicts characteristics of a beta-sheet domains (amino acid residues 6–14) and an alpha-helix (amino acid residues 19–24) based on bioinformatic analysis. (D, E) 3D structure analyses are performed using the Phyre2 and (http://www.sbg.bio.ic.ac.uk/phyre2/) (D), and AlphaFold web portals (https://alphafold.ebi.ac.uk/) (E).