Fig. 1 |. Generation and testing of Xon.
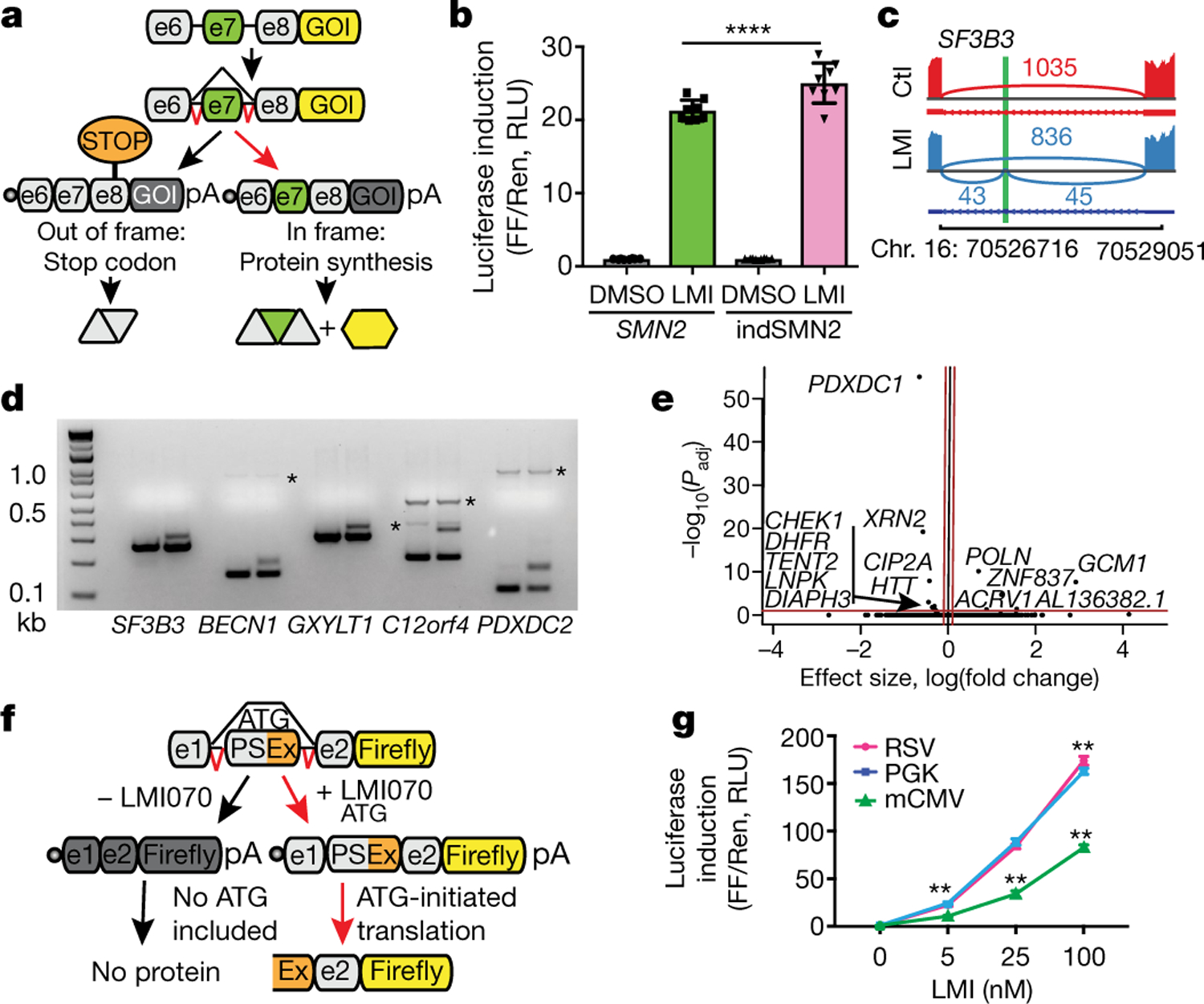
a, Schematic depicting the SMN2-on cassette. In the absence of exon 7 (e7), a premature stop codon blocks translation of the gene of interest (GOI). Inclusion of e7 permits translation. b, Change in firefly (FF) luciferase activity in LMI070-treated (100 nM) compared with DMSO-treated cells, normalized to Renilla (Ren) luciferase activity. RLU, relative light units. Data are mean ± s.e.m. of 8 biological replicates, ****P < 0.0001 SMN2 versus indSMN2, two-way ANOVA, Bonferroni’s post-hoc test. c, SF3B3 splicing without (red; control (Ctl)) or with (blue) 25 nM LMI070 treatment. The genomic location of the LMI070 spliced-in SF3B3 exon (green bar) and intron counts are indicated. d, cDNAs for SF3B3, BECN1, GXYLT1, C12orf4 and PDXDC2 amplified from cells treated with DMSO or LMI070 (25 nM) (one representative plot from 4 RNA-seq samples). Novel splicing was confirmed by Sanger sequencing. The asterisks represent non-specific bands. e, Volcano plot illustrating differentially expressed genes between DMSO-treated and LMI070- treated cells. The red horizontal bar represents 0.05 significance on a −log10 scale. The red vertical bars indicate thresholds of −0.1 and 0.1 fold change. Genes meeting the significance and minimum fold-change thresholds are labelled (4 samples per group). f, Candidate minigenes for translation control; e1 and e2 are the flanking exons of the novel SF3B3 exon (Extended Data Tables 1, 2). g, Induction of luciferase activity in cells transfected with the SF3B3-based Xon cassette. Samples are normalized to Renilla luciferase and are relative to DMSO. Data are mean ± s.e.m. of 8 biological replicates, **P < 0.01 mCMV versus RSV or PGK; **P < 0.01 RSV versus PGK, two-way ANOVA followed by Bonferroni’s post hoc test.
