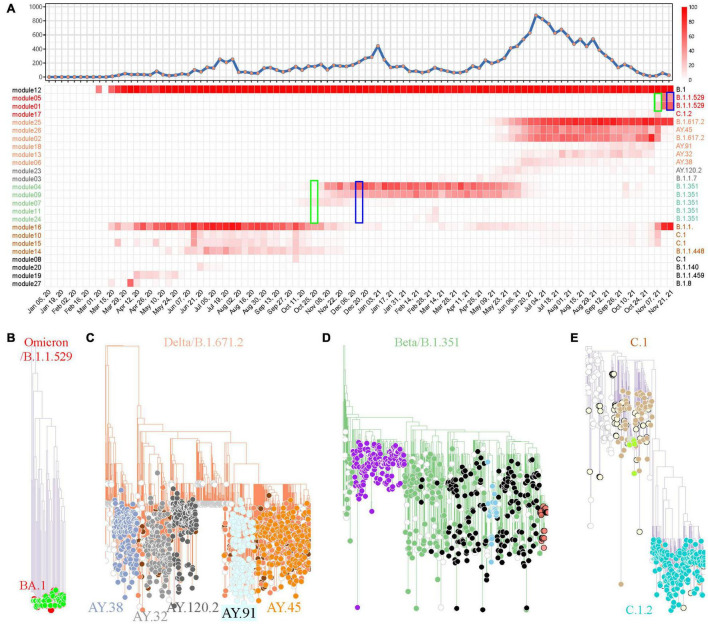
FIGURE 5.
SARS-CoV-2 variant surveillance in South Africa using the weighted network. (A) Weekly distribution of SARS-CoV-2 genome sequences according to sampling time (top) and the heatmap of module-based variant prevalence (bottom). Core mutations within each module were used to define the variants. The modules were reordered and colored according to their module clusters and time course. The weeks when Omicron (B.1.1.529) and Beta (B.1.351) were identified as prevalent variants by network model (green) or reported as VUI or VOC by WHO (blue) are highlighted by rectangles. (B–E) Phylogenic evaluation of every major SARS-CoV-2 variant detected in South Africa, including Omicron (B), Delta (C), Beta (D) and C.1 (E). The module-based variants having consistent classification with Pangolin lineages are labeled.