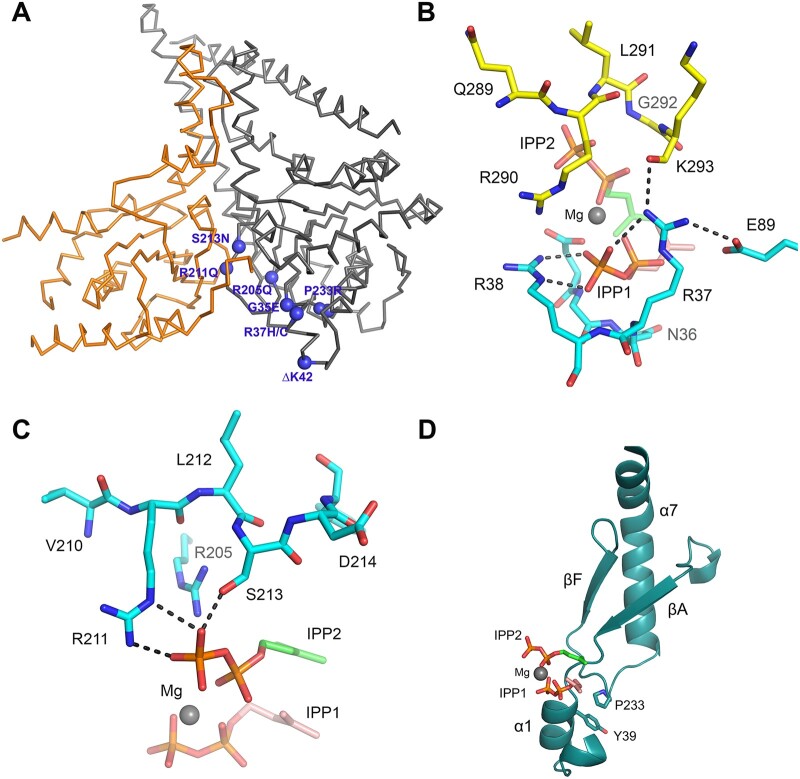
Figure 6.
Mapping of de novo disease-causing DHDDS variants. (A) Ribbon diagram showing the structure of the NgBR/DHDDS complex (adopted from PDB structure 6W2L). DHDDS mutations are indicated as purple spheres on the structure. NgBR is coloured in orange and DHDDS in grey. Most of the mutations cluster around the active site of the complex, including S213N, R211Q, R205Q, G35E, P233R and R37H/C. An exception is I135T, which is distal to the active site. (B) The side chain of R37 is involved in a network of interactions with the pyrophosphate moiety of the allylic substrate, the side chain of a conserved E89 on DHDDS, and the main chain amide of K293 from the NgBR C-terminal region. Hydrogen bonds are shown as black-dashed lines. Nitrogen atoms are in blue, phosphorus atoms are in orange and oxygen atoms are in red. The carbon atoms of the allylic substrate are shown in salmon and those for the homoallylic substrate are shown in green. Mg2+ is shown as a grey sphere. (C) Side chains of R205, R211 and S213 are depicted at the S2 site, and are involved in salt bridging and hydrogen bonding with the pyrophosphate group of the IPP substrate. Replacement of Arg residues with Gln will eliminate these salt bridges and thus weaken IPP binding. Substituting Ser with Asn might result in steric clashes with nearby residues, thereby destabilizing the structure. (D) Ribbon diagram showing the side chain of P233 involved in hydrophobic packing against Y39, which is located at the α1 helix known to harbour important residues for FPP binding. A P233R mutation will disrupt this packing interaction and may destabilize the α1 helix.