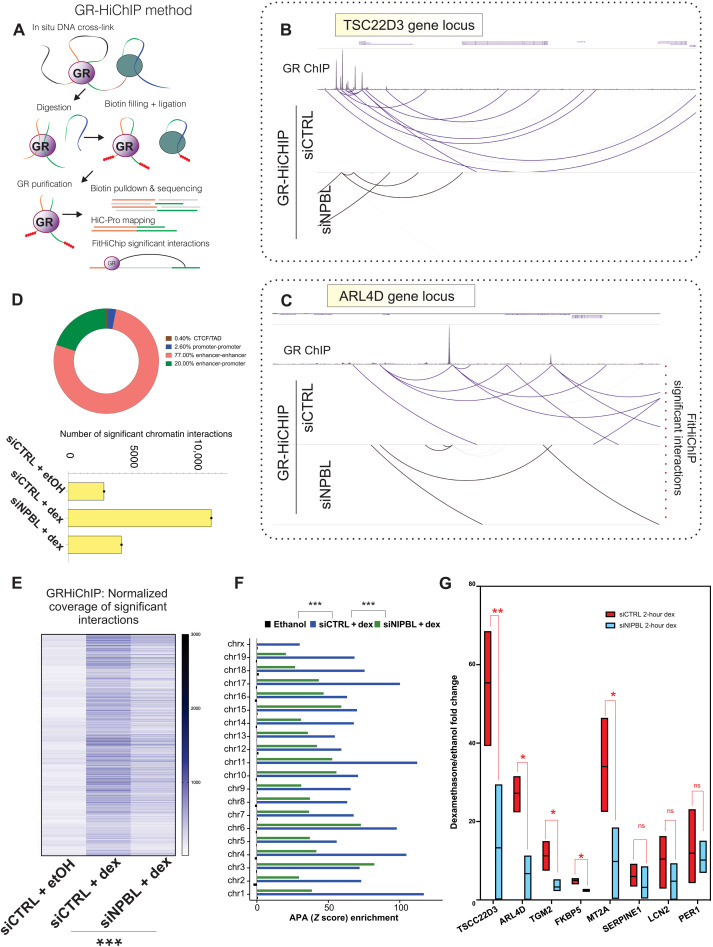
Fig. 2. NIPBL regulates GR-bound long-range interactions and gene regulation.
(A) Experimental design of the GR-HiChIP protocol and analysis pipeline. (B and C) WashU genome browser screenshot of GR-bound long-range interaction identified by FitHiChIP at the TSC22D3 and ARL4D locations, in siCTRL and siNIPBL cells. Loop strength is normalized at a 5-kb resolution. (D) Top: The number of significant GR long-range interactions identified by FitHiChIP, at a Q value of 0.01 using coverage bias parameters, in untreated cells, siCTRL + 1-hour dexamethasone, and siNIPBL + 1-hour dexamethasone. Bottom: The percentages of the GR-bond chromatin interactions stratified into different types of long-range interactions. (E) Quantification intensity of the commonly identified GR-HiChIP long-range interactions identified by FitHiChIP (Q value of 0.01) in siCTRL and siNIPBL samples, at a 5-kb resolution. NIPBL depletion leads to profound reduction of GR-bound chromatin interaction number and looping strength. P < 0.00001 derived from KS test. (F) Aggregate enrichment for each mouse chromosome in the GR-HiChIP datasets in EtOH, siCTRL, and siNIPBL samples. Aggregate peak analysis (APA) measurements (Z score) were calculated on the long-range interactions identified by FitHiChIP (Q value of 0.01) using the juicertools at a 5-kb resolution and VC normalization. P < 0.00001 derived from KS test. (G) Nascent RNA quantification, measured by qRT-PCR, of GR target genes (1 hour of EtOH or dexamethasone treatment) after 48 hours of siRNA transfection against nontargeting sequence (siCTRL) or NIPBL. Fold inductions (dexamethasone/EtOH) are normalized on glyceraldehyde-3-phosphate dehydrogenase (GAPDH) nascent RNA. Error bars represent SE, and P value was derived from unpaired Mann-Whitney tests (n = 3 independent biological replicates). ns, not significant.