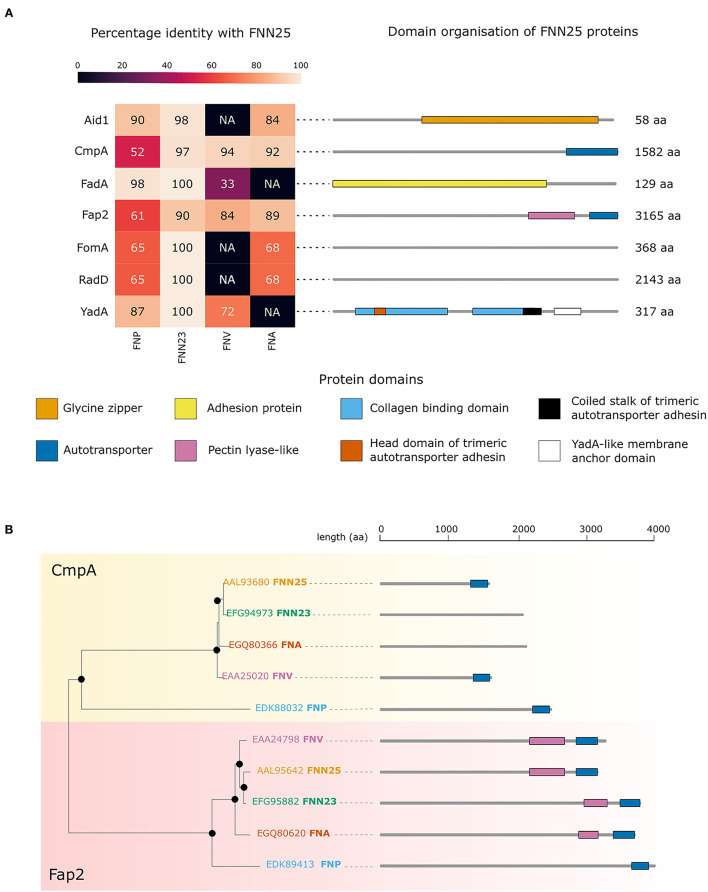
Figure 4.
Bioinformatic analysis of adhesion proteins in F. nucleatum subspecies. (A) Conservation of adhesion proteins in ATCC strains of F. nucleatum. The heatmap shows the percentage of identity with the FNN25 adhesion proteins Aid1, CmpA, FadA, Fap2, FomA, RadD, YadA. Protein length and domain organisation are indicated on the right for each of FNN25 proteins. The domains are indicated and coloured differently: glycine zipper (orange); autotransporter (dark blue); adhesion protein (yellow); pectin lyase-like (pink); collagen binding domain (light blue); head domain of trimeric autotransporter adhesin (dark orange); coiled stalk of trimeric autotransporter adhesin (black); YadA-like membrane anchor domain (white). (B) CmpA and Fap2 phylogenetic tree. The CmpA branch is highlighted in yellow, the Fap2 branch is highlighted in pink. Black circles represent bootstrap values > 95. FNN25 proteins are coloured in orange; FNN23 proteins in green; FNA proteins in dark orange; FNV in pink; FNP in light blue. Protein structures and lengths are outlined on the right of the tree. Autotransporter domains are coloured in blue, pectin lyase-like domains in pink. Figures were drawn with seaborn (version 0.11.2) and R (version 4.1.2).