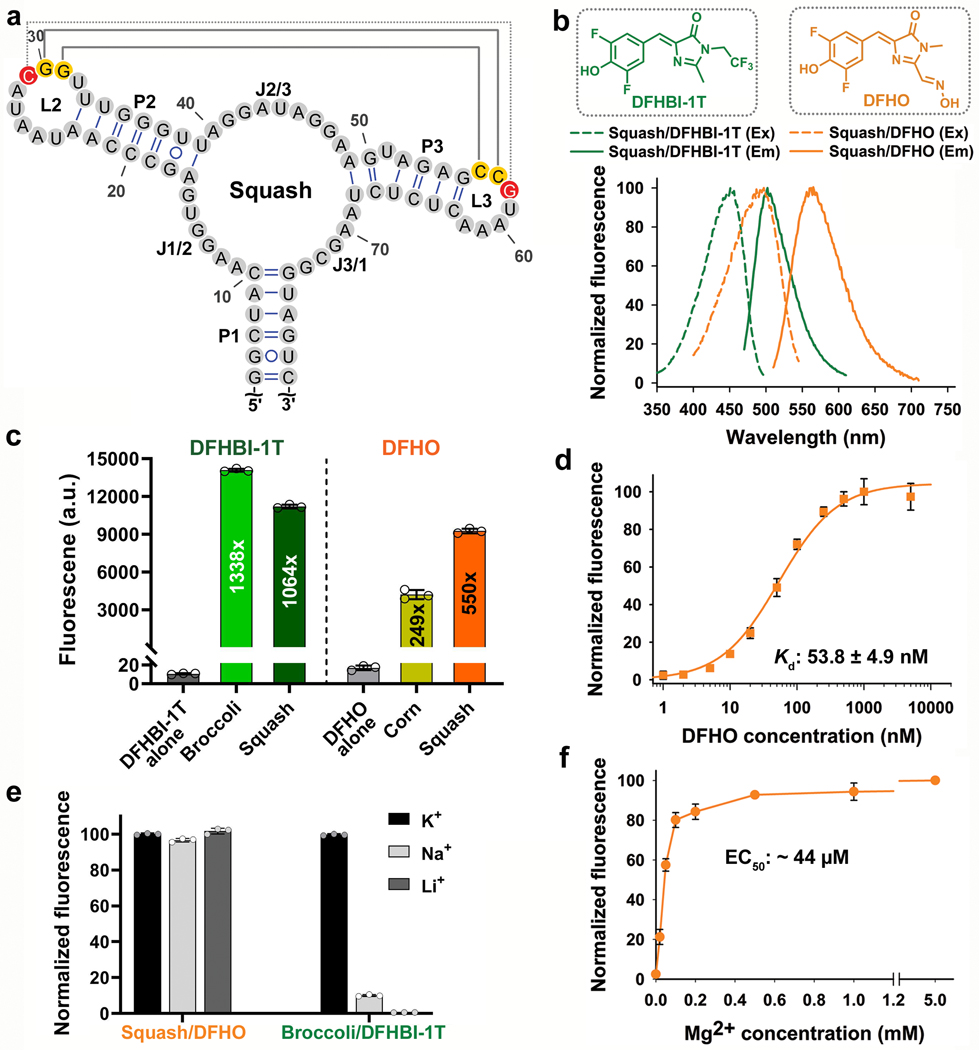
Fig. 2 |. Squash activates the fluorescence of DFHBI-1T and DFHO without utilizing a G-quadruplex.
a, Secondary structure of Squash as predicted by mFOLD. Squash retained the main structural elements of add A-riboswitch aptamer including a 3-way junction and predicted kissing loop interactions (yellow residues, solid gray line). An additional basepair in the kissing loop between loop L2 and L3 (red residues, dotted gray line) improved fluorescence activation. b, Squash binds and activates the fluorescence of both DFHBI-1T and DFHO. Shown are the excitation (Ex) and emission (Em) spectra of Squash bound to DFHBI-1T and DFHO (structures shown in inset). Spectra were measured using 20 μM RNA and 2 μM of the indicated fluorogenic dye. c, Squash shows similar fluorescence activation of DFHBI-1T as Broccoli but much higher activation of DFHO than Corn. Fluorescence activation was measured by incubating 200 nM dye and 10 μM RNA. By using a large excess of RNA compared to the fluorophore, we ensured that the fluorescence observed is from 200 nM RNA-fluorophore complex. Data represent mean values ± s.d. for n=3 independent experiments. The values inside the bars indicate fold activation. AU, arbitrary units. d, The dissociation constant (Kd) between Squash and DFHO was measured by titration of 50 nM Squash with increasing concentration of DFHO, and then the data were fitted using a one-site saturation model. Data represent mean values ± s.d. for n=3 independent experiments. e, Squash-DFHO fluorescence was measured in buffers containing exclusively the indicated cations. Squash was fluorescent in K+-free buffers suggesting that it lacks a G-quadruplex. Broccoli, which contains a G-quadruplex, exhibited markedly reduced fluorescence in the absence of K+. Data represent mean values ± s.d. for n=3 independent experiments. f, Squash-DFHO shows >80% fluorescence activation at 0.1 mM MgCl2. Similar to the parental add A-riboswitch aptamer (see ref. 16), Squash-DFHO has a low magnesium requirement for fluorescence activation.. Data represent mean values ± s.d. for n=3 independent experiments.