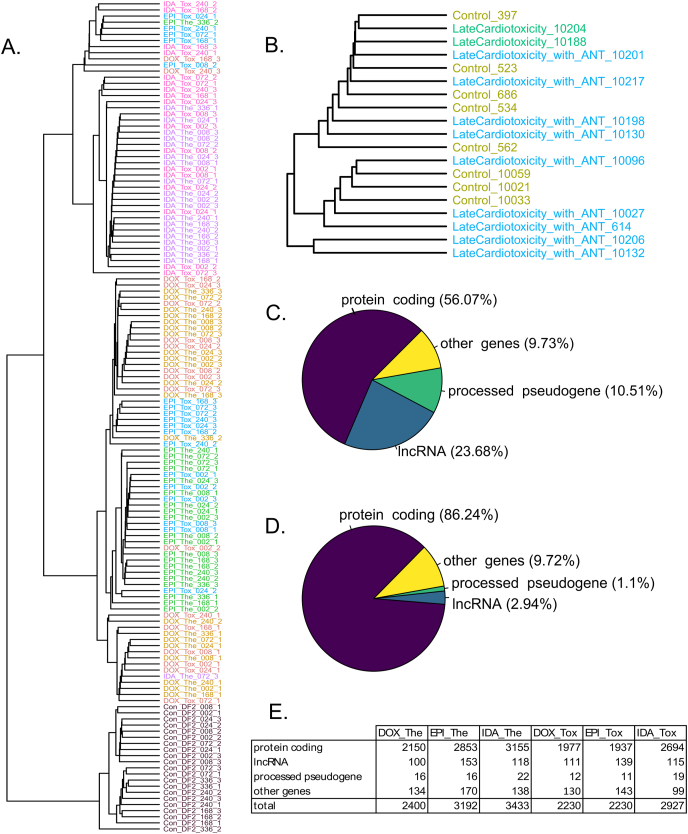
Fig. 1.
A general view of transcriptome profiles of the in vitro and biopsy samples. (A) The cluster tree of the in vitro samples' transcriptome profiles. (B) The cluster tree of the biopsy samples' transcriptome profiles. The biopsy samples included heart failure patients without a cancer history (Control) and heart failure patients who underwent cancer treatment with ANT (LateCardiotoxicity_with_ANT) and without ANT (LateCardiotoxicity). The ending numbers are patient IDs (C) The gene type of expressed genes (the average read count across samples >0, annotated using the Ensembl database) in the in vitro samples. (D) Gene types of overlapped differentially expressed genes (545 genes) across all in vitro ANT-treated conditions compared to control samples. (E) The number of differentially expressed genes in each in vitro ANT-treated condition compared to control samples. P-adj <0.01. Con_DF2: control samples; DOX: doxorubicin, EPI: epirubicin, IDA: idarubicin; The: therapeutic dose, Tox: toxic dose; 002, 008, 024, 072, 168, 240, 336 are corresponding exposure periods; ANT: anthracycline(s).