Fig. 4 |. Deep mutational scanning and CRISPR/Cas9 directed editing of binding site residues reveals mutations which confer resistance to ammocidin and apoptolidin.
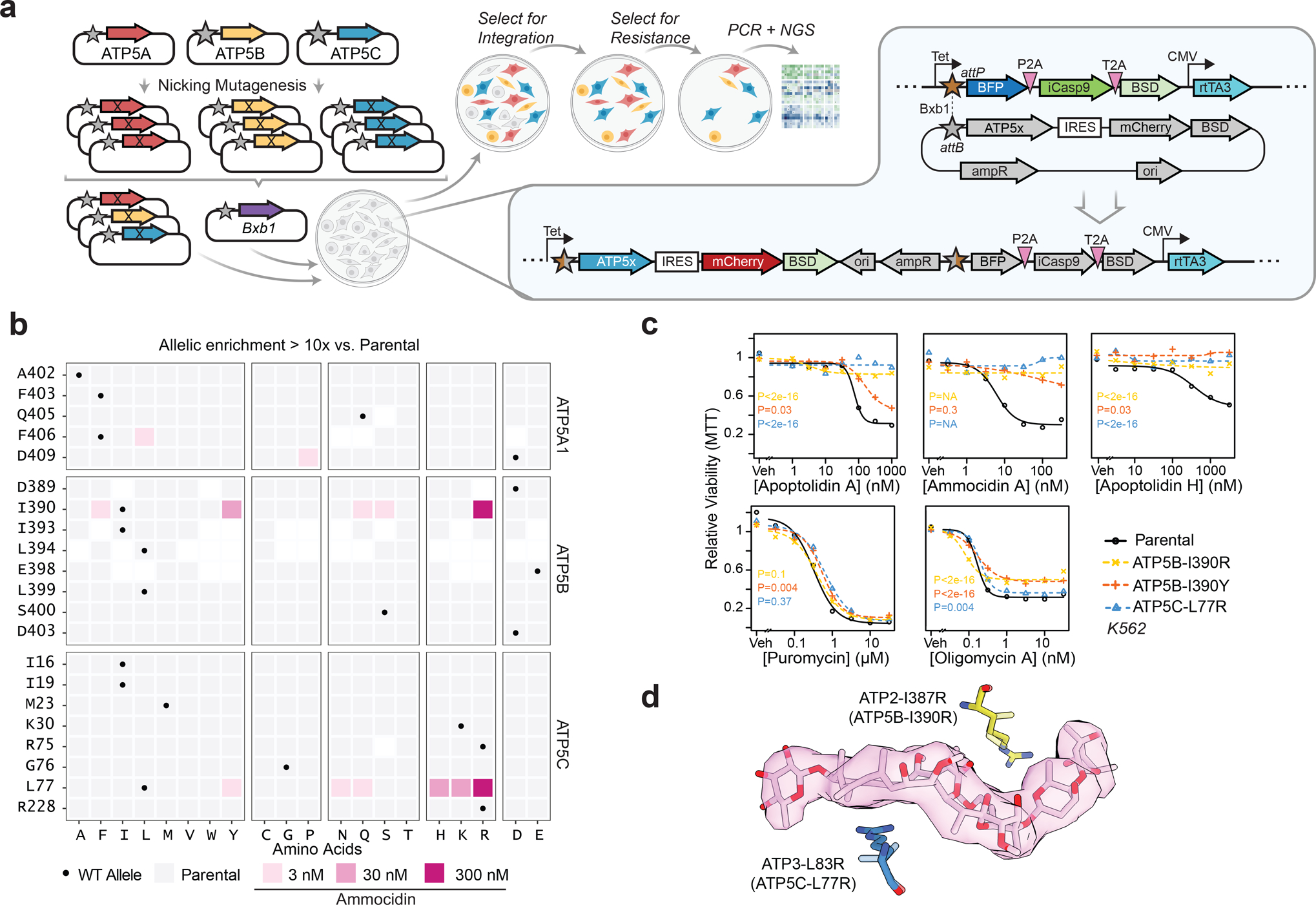
a, Illustration of the Bxb1 landing pad system consisting of an attP containing landing pad integrated at a single site in the genome and an attB transfer plasmid containing the variant gene of interest (ATP5x), colored genes are expressed in the presence of doxycycline; b, Tile plot of allelic enrichment after selection, filled based on the highest dose at which >10-fold enrichment was observed; c, Comparison of drug sensitivity between K562 Parental and K562 CRISPR/Cas9 knock-in resistant mutants demonstrating cross resistance to glycomacrolides with retained sensitivity to Oligm and puromycin (n = 3 per dose, P values computed for lower-bound of fitted log-logistic model, two-sided); d, Modeling of L83R and I387R mutations in the ammocidin cryoEM model (~ human residues in parenthesis).
