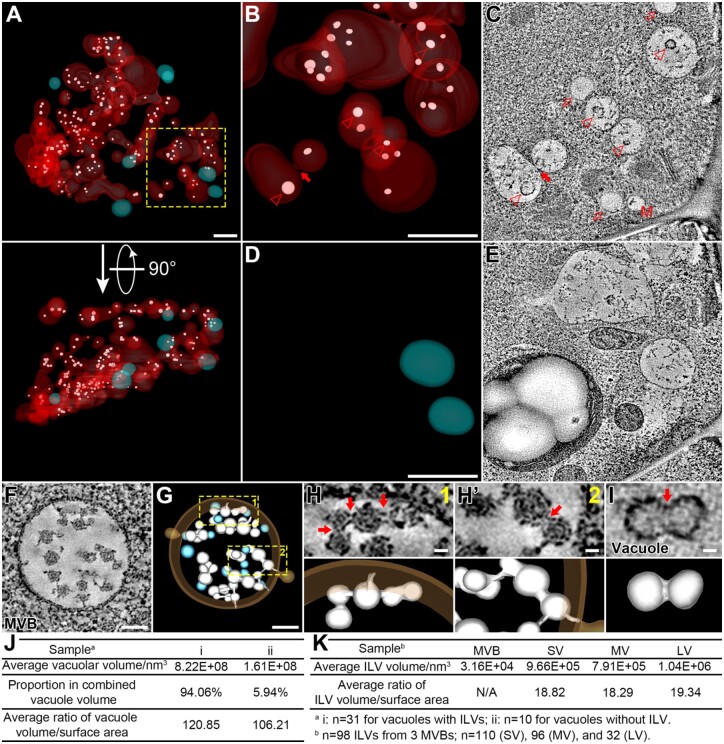
Figure 4.
Most vacuoles in GMC contain swollen ILVs. A, The whole-cell ET model of vacuoles and swollen ILVs in GMC. 3D models are color-coded: Vacuoles with ILVs (red), vacuoles without ILV (cyan), and ILVs (white). All 3D models are shown in semi-transparent fashion. A perpendicular view is shown in bottom part. Scale bar, 1,000 nm. Also see Supplemental Movie S7. B–E, The indicated square box in (A) was further zoomed-in for ET analysis with enlarged models shown in (B) and (D). Vacuoles without ILV were omitted in (B) and vacuoles with ILVs were omitted in (D). Their representative tomographic slices are shown in (C) and (E), respectively. Arrowheads indicate swollen ILVs in tomographic slice (C) and their corresponding 3D models (B). The closed arrow indicates a direct membrane connection between two vacuolar compartments with ILVs (B and C). Open arrows indicate lipid droplets (C). M, multivesicular body, MVB. Scale bars, 1,000 nm for all panels. F and G, Representative tomographic slice (F) and corresponding 3D model (G) of a MVB from WT Arabidopsis cotyledon epidermis. Single ILVs were colored in cyan and concatenated ILVs were colored in white. WT, wild-type. Scale bars, 100 nm. H and H′, The indicated square boxes in (G) were further zoomed-in, showing details of the concatenated ILV buds connected to the endosomal limiting membrane. Red arrows (upper) indicate membrane interconnections with corresponding 3D models (lower). Scale bars, 20 nm for all figure parts. I, Representative tomographic slice (upper) and corresponding 3D model (lower) of a swollen ILV in GMC. Red arrow indicates shallow invagination on the membrane surface. Scale bar, 20 nm. J, Quantification analyses on vacuolar parameters of the two types of vacuoles in GMC: i, vacuoles with ILVs and ii, vacuoles without ILV. N = 31 for vacuoles with ILVs, n = 10 for vacuoles without ILV. K, Quantification analyses on ILV parameters: n = 98 ILVs from 3 MVBs; n = 110 (SV), 96 (MV), and 32 (LV). SV, small vacuole; MV, medium vacuole; LV, large vacuole.