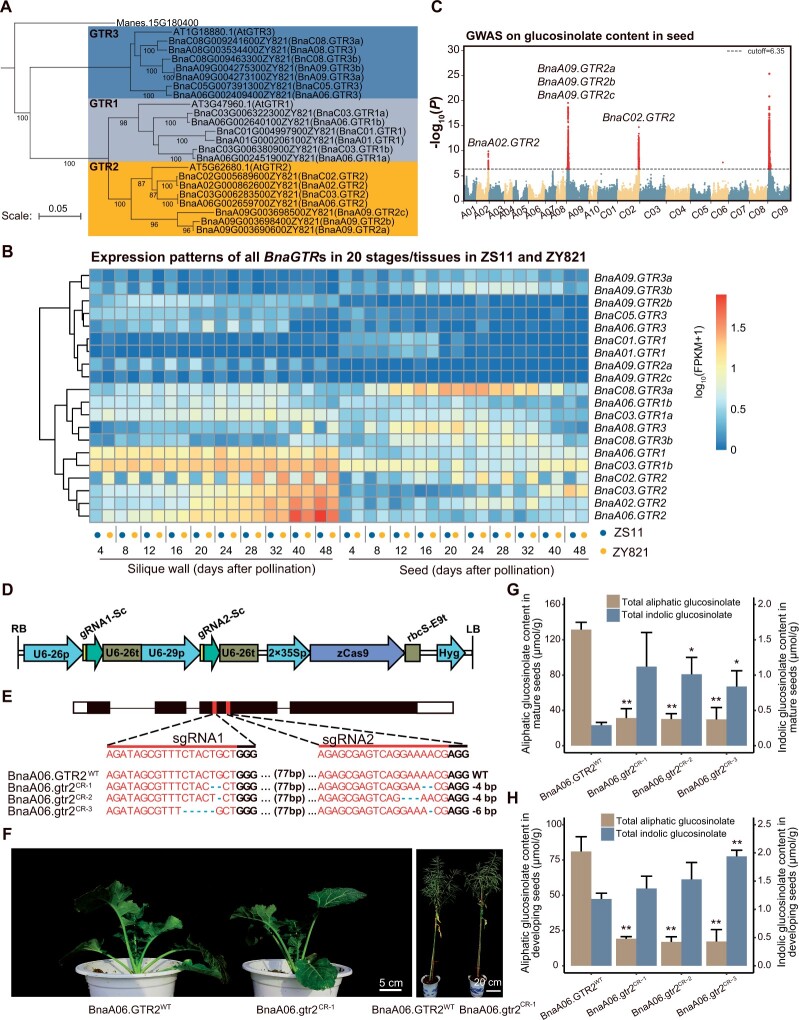
Figure 1.
Identification of a glucosinolate transporter gene with rare natural mutation in B. napus. A, Neighbor-joining tree of GTRs homologs in A. thaliana and B. napus. MANIHOT ESCULENTA CYANOGENIC GLUCOSIDE TRANSPORTER-1 (MeCGTR1) was used as an out-group. Bootstrap values above 75% were given at branch nodes. The scale bar indicates the number of amino acid residue substitutions per site. B, Expression patterns of BnaGTR2s in the low (ZS11) and high (ZY821) seed glucosinolate accessions in the different tissues at the different growth stages. C, Manhattan plot of genome-wide association study on seed glucosinolate content. Horizontal dashed line represents the significant threshold (−log10(P) = 6.35). Red dots indicated the significantly associated single-nucleotide polymorphisms with seed glucosinolate content. D, Schematic of the constructed genome-editing vector with two gRNAs to target BnaA06.GTR2 sequence. RB/LB, right/left border of T-DNA; U6-26p and U6-29p, two A. thaliana U6 gene promoters; 35Sp, cauliflower mosaic virus 35S promoter; U6-26t, A. thaliana U6 gene terminator; rbcS-E9t, rbcS-E9 gene terminator; gRNA1-Sc and gRNA2-Sc, guide RNA scaffold; zCas9, Zea mays codon-optimized Cas9; Hyg, hygromycin resistance gene. E, Characterization of edited BnaA06.GTR2 transgenic lines. The upper panel shows gene structure and CRISPR–Cas9 target sites in BnaA06.GTR2. The Open boxes indicate 5'-untranslated region (5'-UTR) or 3′-UTR; the black boxes indicate exons; the black lines indicate the introns; The red boxes indicate sgRNA target sites. The lower panel shows the sequences of three T0 independent transgenic lines in the target sites. Deletions are indicated as blue hyphens. F, The phenotype of BnaA06.gtr2CR-1 T2 mutant during the vegetative (left) and reproductive (right) stages. G and H, Glucosinolate content of BnaA06.GTR2 knockout mutants and the transgene-negative control (BnaA06.GTR2WT) in mature seeds (G) and developing seeds at 40 d after pollination (H). In (G) and (H), at least four plants per line were independently sampled for measurement. Each error bar is means ± sd (n ≥ 4 plants for each line). Student’s t test was used for statistical analysis, single asterisk indicates significant differences at P < 0.05, double asterisk indicates significant differences at P < 0.01, all compared to the BnaA06.GTR2WT.