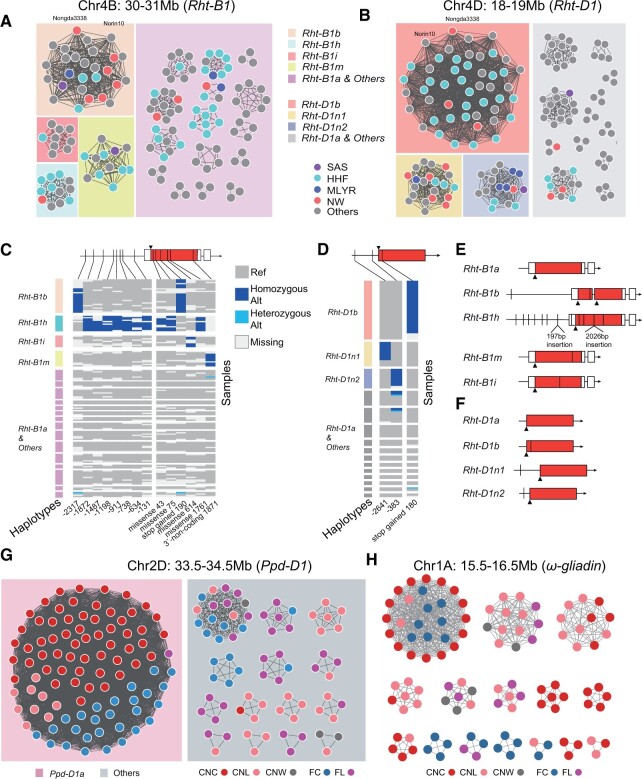
Figure 5.
Local-scale GGNet helps reveal the selection of gene located blocks during breeding. GGNet of the genomic windows with Rht-B1 (A) and Rht-D1 (B). Each node represents an accession. Node colors, different agro-ecological zones. SAS, Southwestern autumn-sown spring wheat zone. HHF, Huang Huai facultative wheat zone. MLYR, Middle and lower Yangtze River valley autumn-sown spring wheat zone. NW, Northern winter wheat zone. An edge indicates that the two connected nodes share the same gHap. The rectangle background colors indicate major selected clusters and others, with the corresponding alleles labeled in the squared legend. C and D, The heatmap shows sequence variations with groups corresponding to alleles of Rht-B1 (C) and Rht-D1 (D). Dark blue, homozygous variation. Light blue, heterozygous variation. Gray, genotype of the reference genome (CS). White, missing data. The variation positions at the bottom are the relative distances from the start codon. In the left annotation track, the group colors are consistent with the box colors in (A) and (B). The triangles represent start codon positions. Red region in boxes, coding regions. White region in boxes, Untranslated Region (UTR) sequences. E and F, Gene structure features of different alleles of Rht-B1 (E) and Rht-D1 (F). Rht-D1n1 and Rht-D1n2 are two unreported alleles found in this study. GGNets of the genomic windows with Ppd-D1 (G) and ω-gliadin (H). Each node represents an accession. Node colors, CNC, CNL, and CNW and foreign cultivars and landraces. An edge indicates that the two connected nodes share the same gHap. The rectangle background colors in (G) indicate whether accessions carrying Ppd-D1a or not.