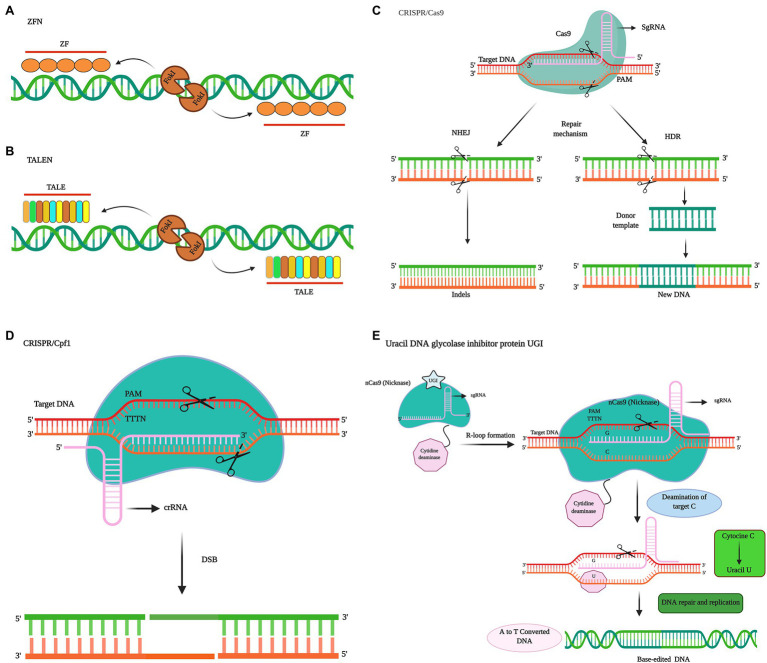
Figure 1.
Comparison of different widely used genome editing systems in plants. Site-specific techniques, such as those based on zinc finger nucleases (ZFNs; A) and protein-dependent DNA cleavage systems, such as those based on transcription activator-like effector nucleases (TALENs; B). The two sequence-specific zinc finger proteins flank the target sequence, and FokI nuclease follows the C terminus of each protein. Zinc finger proteins are used to direct FokI dimers to target the specific DNA sites to be cleaved. TALENs consist of two sequence-specific TALEN proteins arranged on the target sequence, with FokI nuclease followed by the C terminus of each protein. The TALEN protein directs FokI dimers to the target specific DNA sites to be cleaved. (C) Illustration of a cluster of the regularly spaced short palindrome repeats (CRISPR)/CRISPR-associated nuclease 9 (Cas9) system. CRISPR-Cas induces double-strand breaks (DSB) in the DNA strand. CRISPR RNA (crRNA) directs the Cas9 protein. Trans-activating CRISPR RNA (tracrRNA) is used to stabilize the structure and activate Cas9 to cut the target DNA. Single guide RNA [sgRNA (Gulabi)] recognizes the target gene, and then the Cas9 protein (green) cuts the two strands of the target DNA at the RuvC and His Asn-His (HNH) domains. The cutting of DNA by Cas9 is dependent on the presence of spacer adjacent motif (PAM) sequences. A 20-base sgRNA defines the binding specificity. In DNA, there are two mechanism of double-strand break (DSB) repair, namely, homology-directed repair (HDR), which is activated in the presence of a template and leads to knock-in or gene replacement, and non-homologous end joining (NHEJ), which is not sufficiently precise and leads to permanent gene knockout. (D) The Cpf1 system. Cpf1 is a two-component RNA programmable DNA nuclease related to CRISPR. The PAM of Cpf1 is TTTN. Cpf1 cuts the target DNA and introduces a DSB, a 5-nt staggered cut at the 5′ end of the T-rich PAM. CRISPR-Cas9 nickase introduces breaks only in the strand complementary to sgRNA. The double nickase with two sgRNAs introduces a staggered DSB into the DNA, after which HDR repairs the DSB. Uracil DNA glycosylase inhibitor (UGI) protein prevents removal of uracil from the DNA and subsequent repair pathways and contributes to increasing the frequency of mutations.