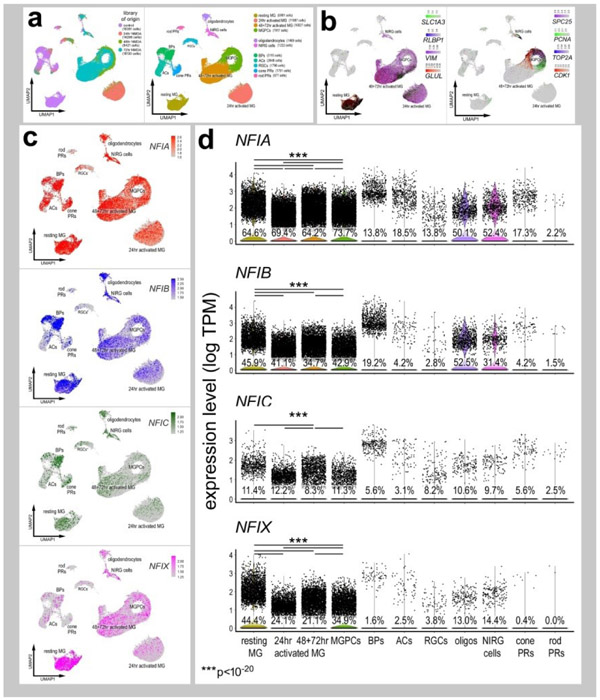
Figure 2. Patterns of expression of NFIA, NFIB, NFIC and NFIX in normal and NMDA-damaged chick retinas.
scRNA-seq was used to identify patterns of expression of NFI’s among retinal cells with the data presented in UMAP (a-c) or violin plots (d). scRNA-seq libraries were aggregated for retinal cells from control eyes and eyes 24hr, 48hr, and 72hr after NMDA-treatment (a). UMAP-ordered cells formed distinct clusters of neuronal cells, resting MG, early activated MG, activated MG and MGPCs (a,b). Resting MG were identified based on elevated expression of SLC1A3, RLBP1, VIM and GLUL (b). Activated MG down-regulated SLC1A3, RLBP1 and GLUL, and MGPCs up-regulated proliferation markers including SPC25, PCNA, TOP2A and CDK1 (b). UMAP heatmaps of NFIA, NFIB, NFIC and NFIX demonstrate patterns of expression across retinal cell types. Violin plots illustrate the percentage of expressing cells, levels of gene expression and significant changes (***p<10exp-20) in levels (log TPM) were determined by using a Wilcox rank sum with Bonferoni correction.