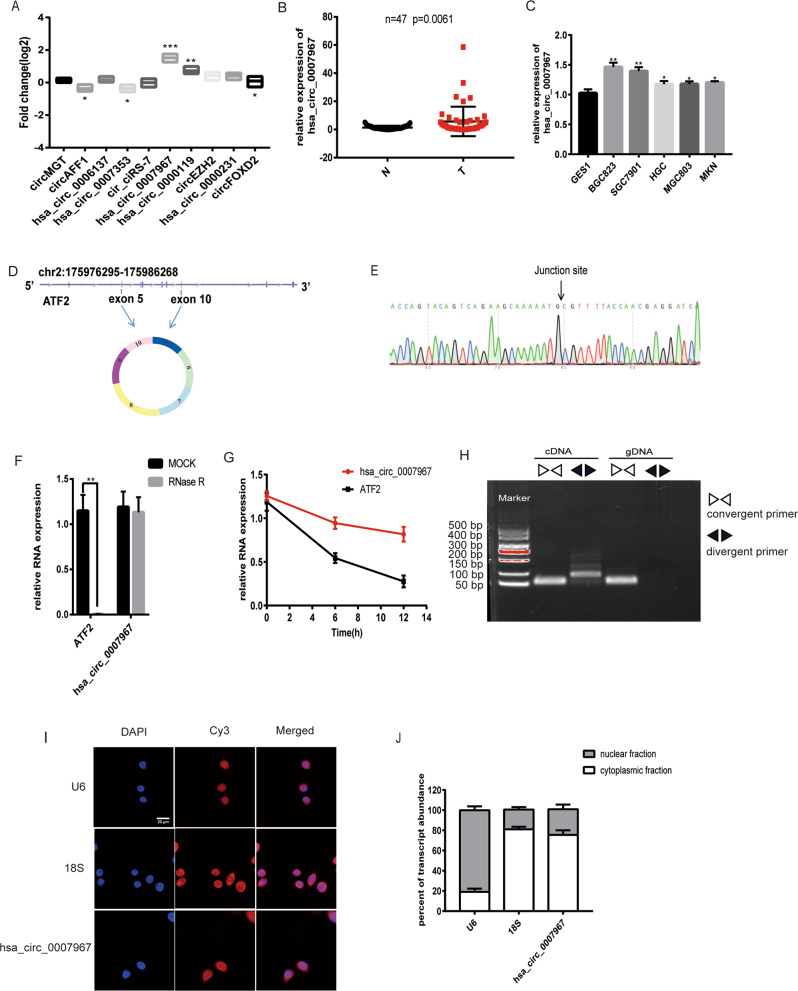
Fig. 1. Identification and characterization of hsa_circ_0007967.
A Expression of five most upregulated and downregulated circRNAs in five paired GC tumors and normal tissues. B Expression of hsa_circ_0007967 in 47 pairs of GC tissues and normal tissues. C Expression of hsa_circ_0007967 in GC cell lines and normal gastric epithelial cell line. D The location of hsa_circ_0007967 in chromatin and its structure. E Sanger sequencing confirming the back splicing junction site. F The RNase R digestion assay suggesting hsa_circ_0007967 is resistant to RNase R. G The use of act-D (1 μg/μl) demonstrating that hsa_circ_0007967 is stable than its linear form. H Northern blotting of hsa_circ_0007967 and its linear form in cDNA and gDNA amplified by convergent and divergent primers. I Location of hsa_circ_0007967 in BGC823 cells detected by FISH. J QPCR analysis of nuclear and cytoplasmic fractions conforming that hsa_circ_0007967 is located in cytoplasm.