Fig. 2.
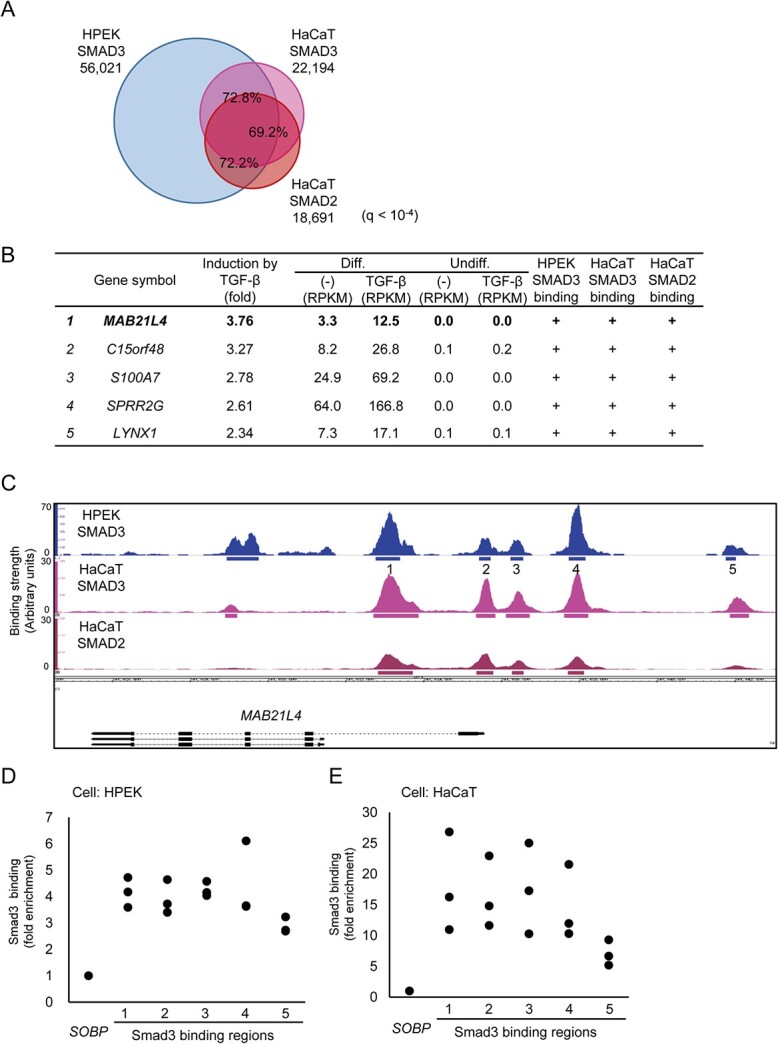
Identification of MAB21L4 as a target of Smad3. (A) Comparison of the Smad3 and Smad2 binding regions in HPEK and HaCaT cells. ChIP-seq data of HaCaT cells stimulated with 1 ng/ml of TGF-β3 for 1.5 h were obtained for comparison to the data obtained in Fig. 1. Significant binding regions were determined at q < 10−4 and overlap analysis was performed by using bedtools (https://bedtools.readthedocs.io/en/latest/). (B) MAB21L4 was identified as the most up-regulated target gene of TGF-β and Smad3 in differentiated HPEK cells. Genes with expression levels of at least 10 FPKM in any of the differentiated HPEK samples were included in the analysis. The data were then sorted by the order of induction by TGF-β. Genes exclusively expressed in the differentiated HPEK cells (FPKM < 1 in undifferentiated HPEK cells) are shown. ‘Smad3 binding’ is defined as significant Smad3 binding within 100 kb from the gene. (C) Smad3 and Smad2 binding regions at the MAB21L4 locus are shown. There are three variants (a longer form and lower two shorter forms) of MAB21L4. Numbers show the regions analysed by ChIP-qPCR in (D) and (E). (D) ChIP-qPCR analysis of Smad3 binding at the MAB21L4 locus. Differentiated HPEK cells were treated with TGF-β for 1.5 h and fixed. The SOBP locus was used as the negative control region (9). Data were obtained from three biological replicates. (E) ChIP-qPCR analysis of Smad3 binding in an epidermal keratinocyte cell line HaCaT. ChIP was performed as in (D). Data were obtained from three biological replicates.
