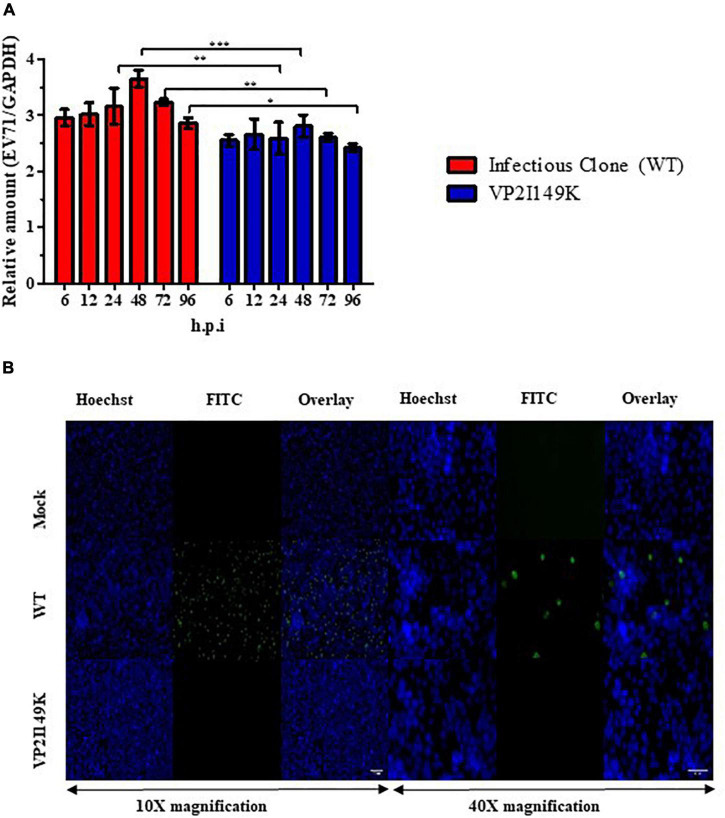
FIGURE 2.
Replication of VP2 I149K mutant in NSC-34 cells. (A) Quantification of intracellular viral RNA. NSC-34 cells were infected with VP2 I149K mutant or WT S41 strain at MOI of 10. At the indicated time-points, infected cells were harvested and viral RNA was extracted and quantified by RT-qPCR. Intracellular viral RNA levels (Ct-values) were normalized to housekeeping GAPDH RNA levels. The fold change between infected vs. uninfected samples was calculated and data were expressed as the mean ± SD of technical triplicates. A two-way ANOVA with Bonferroni correction was used to evaluate statistical significance. *p < 0.05, **p < 0.01, *** p < 0.001. One representative set of two independent experiments is shown. (B) Detection of dsRNA. NSC-34 cells were infected with WT S41 or VP2 I149K mutant at MOI of 10, or left uninfected (mock control). At 72 h.p.i, the cells were fixed and processed for immunostaining using antibodies specific to dsRNA (FITC green). Cell nuclei were stained with Hoechst 33342. The images were captured at 10× or 40× magnification. Representative images are shown. Scale bar denotes 100 and 40 μm for 10× or 40× magnifications, respectively.