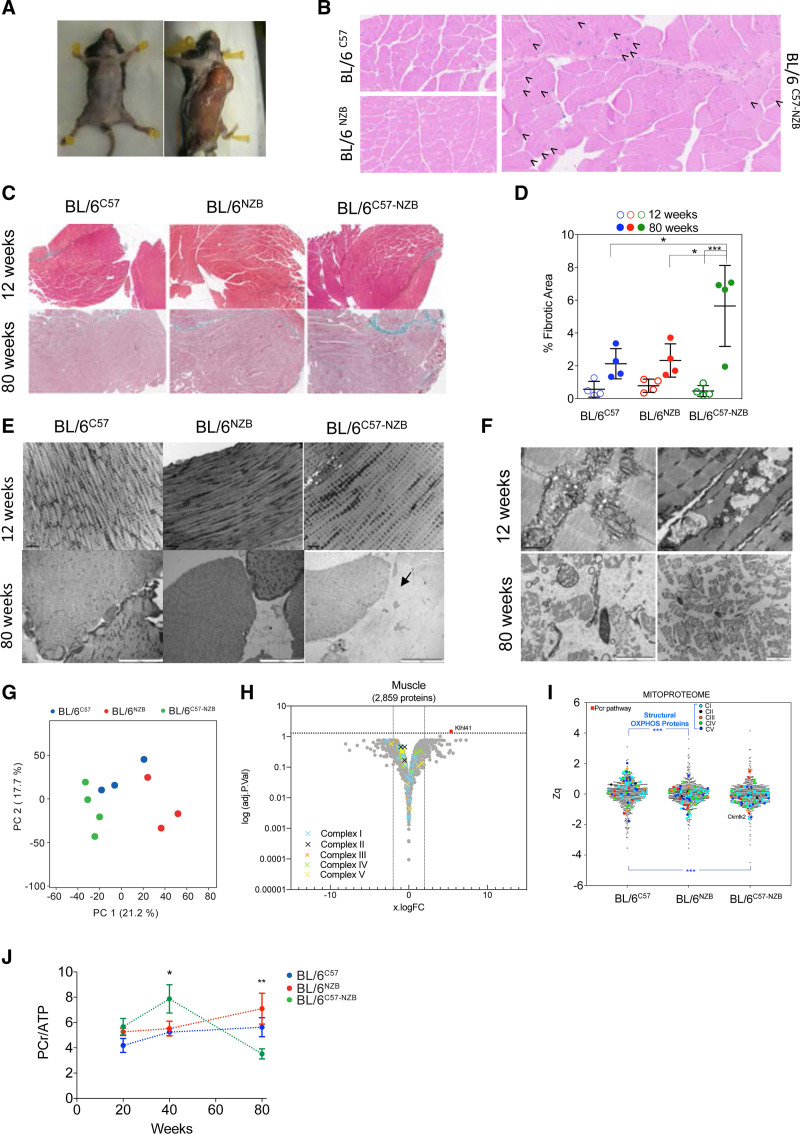
Figure 6.
Analysis of skeletal muscle. A, Representative images showing the loss of muscle mass in heteroplasmic animals. B, Representative hematoxylin and eosin staining on skeletal muscle sections from 80-week-old homoplasmic and heteroplasmic mice. Arrows indicate central nuclei. C and D, Analysis of skeletal muscle collagen and fibrosis in mice of the indicated age and mouse strain. C, Representative images of skeletal muscle histologic sections stained with Masson trichrome staining at the indicated ages and (D) quantification of the fibrotic area. Age (P<0.001) and genotype (P<0.05) as well as their interaction (P<0.01) are significantly different assessed by 2-way analysis of variance. Multiple comparisons corrected by Tukey (n=4 mice per genotype and age). E and F, Electron microscopy analyses of skeletal muscle samples from 12- and 80-week-old mice (3–4 mice per genotype and age). Black arrow indicates a fibrotic area. G through I, Proteomic analysis of skeletal muscle in 12-week-old mice. G, Score plot of the principal component (PC) analysis. H, Volcano plot illustrating proteins with significantly differential abundance between homoplasmic and heteroplasmic gastrocnemius (3 homoplasmic and 4 heteroplasmic samples). The log (adjusted P value) is plotted against the log fold change (logFC; fold change homoplasmic/heteroplasmic animals). On the x axis, vertical lines denote ±2-fold change; on the y axis, the horizontal line denotes the significance threshold (P<0.05 prior to logarithmic transformation). I, MitoProteome analysis highlighting structural oxidative phosphorylation components and mitochondrial elements of the phosphocreatine (PCr) pathway (3–4 mice per genotype). J, 31P magnetic resonance spectroscopy analysis of the PCr/ATP (adenosine triphosphate) ratio in the skeletal muscle (soleus) of different individuals at the indicated age. *P<0.05, **P<0.01, 2-way analysis of variance test. Multiple comparisons corrected by Tukey test (n=5–9). In D and G, dots represent individual mice. In D and J, data are presented as mean±SD. OXPHOS indicates oxidative phosphorylation.