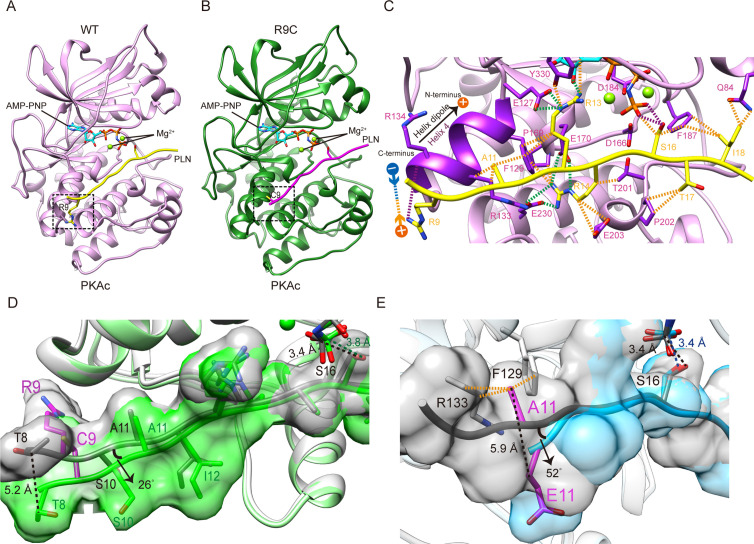
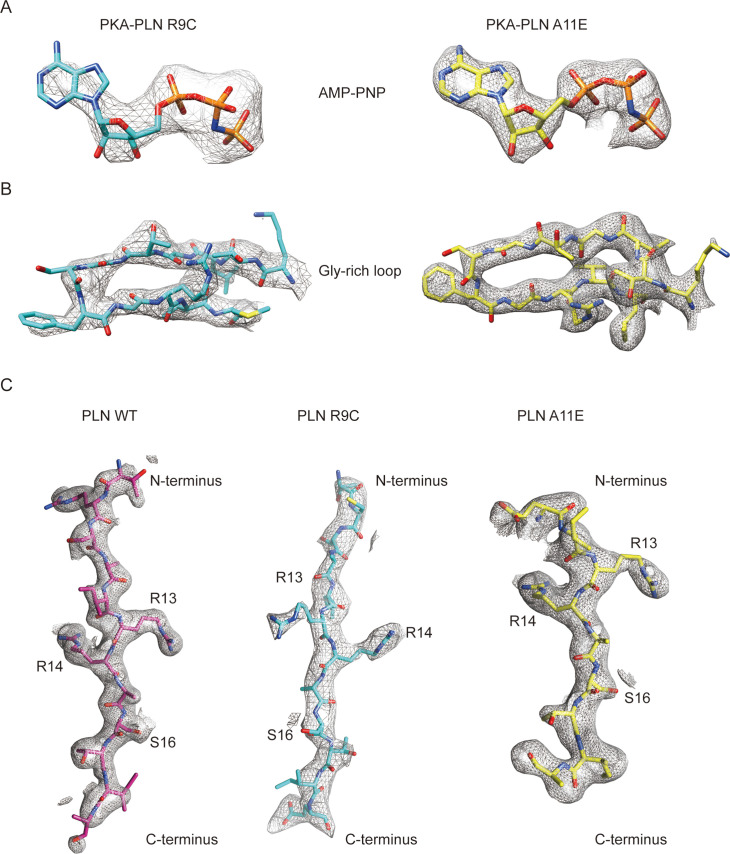
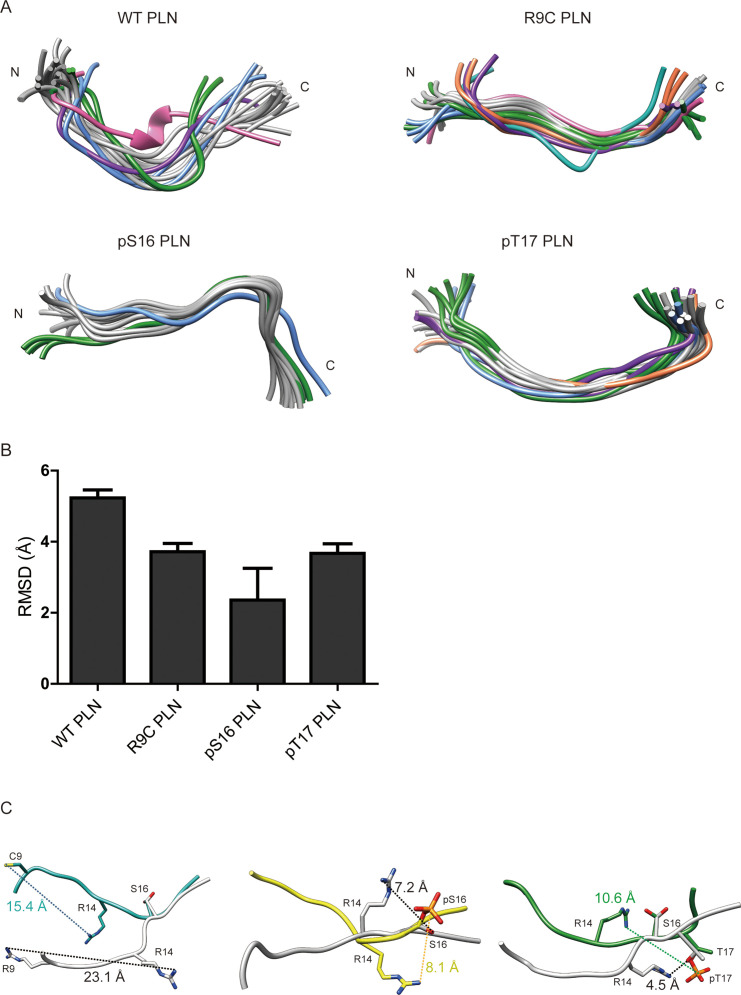
Figure 1. Crystal structures of PKAc-WT/R9C phospholamban (PLN) complex.
(A) Crystal structures of the ternary complex of PKAc, WT PLN, and AMP-PNP. Protein kinase A (PKA) is colored in pink, PLN in yellow, and AMP-PNP in cyan. (B) Crystal structures of the ternary complex of PKAc, R9C PLN, and AMP-PNP. PKA is colored in green, PLN in violet red, and AMP-PNP in cyan. (C) The interaction between PKAc and WT PLN. The van der Waals contacts (orange), the salt bridges (green), and the hydrogen bonds (purple) are indicated by the dash lines. (D) The superposition of PKAc-WT PLN (white-gray) with PKAc-R9C PLN (light green-green). R9C abolishes the electrostatic interaction between Arg9 and the helix dipole of helix 4, inducing conformational changes at the N-terminal region (NTR). (E) The superposition of PKAc:WT PLN (white-gray) with PKAc:PLN A11E (light blue-cyan). A11E forces the NTR to move away from PKAc without affecting the structure at the catalytic center.