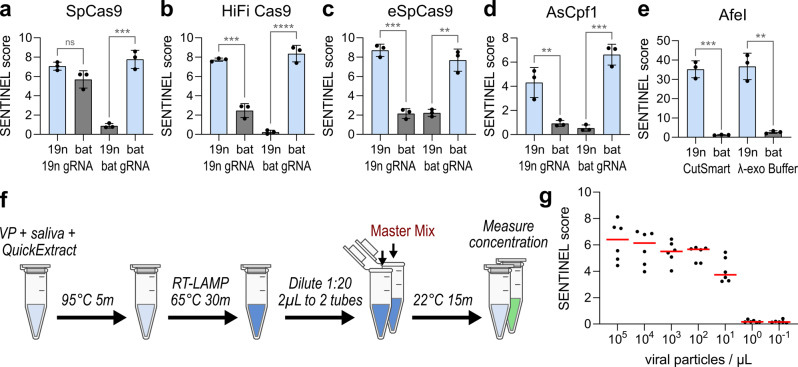
Fig. 4. Compatibility with different sequence-specific endonucleases and with viral particles in human saliva.
a–d Use of SENTINEL to detect the N-gene of SARS-CoV-2 (19n) vs. bat-SL-CoVZC45 (bat), using guide RNA targeting the N-gene of SARS-CoV-2 (19n gRNA) vs. bat-SL-CoVZC45 (bat gRNA). a Wild type SpCas9, b HiFi Cas9, c eSpCas9, and d AsCpf1 were used as the endonuclease. Light blue bars indicate expected positive samples, while gray bars indicate expected negative samples. e Use of AfeI as the endonuclease to discriminate between a single-nucleotide difference in sequence between the N-gene of SARS-CoV-2 (19n) and bat-SL-CoVZC45 (bat). CutSmart, and the λ-exo buffer used with other endonucleases, were both compatible. a–e All error bars represent ±1 standard deviation from mean, from three experimental replicates plotted as round data points. n.s. indicates not significant, * indicates p < 0.05, ** indicates p < 0.01, and *** indicates p < 0.001 from Student’s t-test. f Schematic of SARS-CoV-2 viral particle detection from human saliva using SENTINEL. g Limit of detection from serial dilutions of viral particles in viral transport media, which was mixed into saliva before subject to the SENTINEL assay. Red lines correspond to median of six experimental replicates.