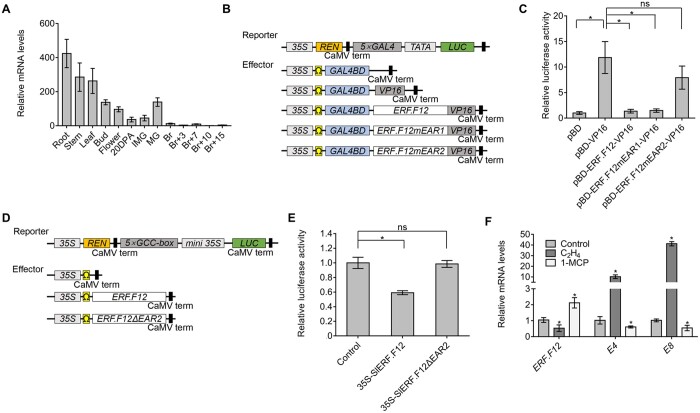
Figure 1.
ERF.F12 displays a ripening-related expression pattern and encodes a transcriptional repressor. A, Relative ERF.F12 transcript levels in different tissues, as assessed by RT-qPCR. 20 DPA, 20 DPA; Br+3–15, 3–15 days after the Br stage. B, Schematic diagram of the double-reporter and effector plasmids in the dual LUC assay for transcriptional inhibition assays. C, Transcriptional repression assays of ERF.F12. The dual LUC/REN reporter was co-transfected with individual effector plasmids into N. benthamiana leaf protoplasts. ERF.F12mEAR1, ERF.F12 with the core Leu residues of EAR1 changed to Ser. ERF.F12mEAR2, ERF.F12 with the core DLN motif of EAR2 changed to SSS. pBD, empty vector, negative control. pBD-VP16, VP16 transcriptional activator domain, positive control. Asterisk indicates statistical significance using Student’s t test, P < 0.05. D, Schematic diagram of the double-reporter and effector plasmids in the dual LUC assay for measuring transcriptional repressor ability of ERF.F12 on a promoter containing the GCC box. E, ERF.F12 represses transcription from a promoter containing a synthetic GCC box. The dual LUC/REN reporter was co-transfected with individual effector plasmids into N. benthamiana leaf protoplasts. ERF.F12ΔEAR2, ERF.F12 with a deletion of EAR2. Asterisk indicates statistical significance using Student’s t test, P < 0.05. F, ERF.F12 transcript levels in WT fruits at the MG stage treated with ethylene (50 μL L−1) for 8 h or 1-MCP (1.0 μL L−1) for 12 h, as determined by RT-qPCR. E4 and E8 are ethylene-responsive marker genes. Asterisks indicate statistical significance using Student’s t test, P < 0.05.